Abstract
Biodegradation of the graphene-based materials is an emerging issue due to their estimated widespread usage in different industries. Indeed, a few concerns have been raised about their biopersistence. Here, we propose the design of surface-functionalized graphene oxide (GO) with the capacity to degrade more effectively compared to unmodified GO using horseradish peroxidase (HRP). For this purpose, we have functionalized the surface of GO with two well-known substrates of HRP namely coumarin and catechol. The biodegradation of all conjugates has been followed by Raman, dynamic light scattering and electron microscopy. Molecular docking and gel electrophoresis have been carried out to gain more insights into the interaction between GO conjugates and HRP. Our studies have revealed better binding when GO is functionalized with coumarin or catechol compared to control GOs. All results prove that GO functionalized with coumarin and catechol moieties display a faster and more efficient biodegradation over GO.
Export citation and abstract BibTeX RIS

Original content from this work may be used under the terms of the Creative Commons Attribution 3.0 licence. Any further distribution of this work must maintain attribution to the author(s) and the title of the work, journal citation and DOI.
Introduction
The adoption of emerging nanomaterials in diverse industrial applications requires understanding and management of their risks for the humans and the environment. Graphene-related nanomaterials (GRMs) are rapidly attracting the attention of many industries including electronics, photonics, optics, composites due to their unique physical and chemical properties [1, 2]. At the same time, their impact on health and environment can also raise concerns [3, 4]. There are several ways how GRMs can cause toxicity, through commercial products, unintentional occupational exposure, and environmental exposure [5]. In this context thorough investigations to know possible risks caused by GRMs are necessary [6].
From the biomedical and environmental point of view, the oxidized form of graphene, graphene oxide (GO), has been proven to be a better candidate over pristine graphene because of its excellent aqueous dispersibility, relative high biocompatibility combined to the remarkable mechanical and optical properties [7]. Consequently, GO has assumed a key role in the biomedical exploration of 2D materials, including the development of new drug delivery systems [8–10], antimicrobial coatings [11], photothermal therapeutics [12], water purification [7], and regenerative medicine [13, 14]. However, the safety profile and biocompatibility of GO in living systems have not been fully assessed and understood yet [15]. For example, it has been reported that a direct injection of highly dispersed GO into lungs resulted in severe and persistent injury along with mitochondrial respiration dysfunction, generation of reactive oxygen species, and activation of inflammatory and apoptotic pathways [16]. Another report showed a predominant retention of GO in lungs of mice for long time inducing pathological changes including inflammatory cell infiltration and granuloma formation [17].
In the context of the above studies, our recent findings on the tissue distribution of highly dispersed GO in mice, reported high renal excretion of GO followed by accumulation of the remaining GO in the reticuloendothelial system organs [18, 19]. The accumulation in these organs was dependent on the thickness of the GO sheets [20]. One recent study also reported a long-term cytocompatibility of aged and fresh GO aqueous suspension [21]. In this scenario, one of the best solutions to reduce the long term impact of GRMs on health and environment is their safe and clean disposal by degradation using natural peroxidases [22].
In the past, it was presumed that biodegradation of carbon nanomaterials was not possible due to their strong resistance and inert graphitic structure. In 2008 Allen et al revealed the biodegradability of oxidized single-walled carbon nanotubes by the plant enzyme horseradish peroxidase [23, 24]. Following this pioneering work, several peroxidases including myeloperoxidase (MPO) [22], eosinophil peroxidase [25], and manganese peroxidase [26] were shown to degrade carbon and other 2D nanomaterials [27, 28]. Similar strategies were then applied to the biodegradation of GO by HRP [29]. Girish et al also demonstrated a possible in vivo degradability of carboxyl-functionalized graphene (treated with nitric acid) in macrophages [30]. Lalwani et al proved that lignin peroxidase is effective in the biodegradation of GO [31]. Liu and co-workers investigated a selective biodegradation of GO covalently functionalized with PEG via a cleavable disulfide bond in the presence of reducing glutathione [32]. More recently, we have highlighted how the hydrophilicity, the percentage of oxygenated groups on the graphitic surface and the aqueous colloidal stability of GO play an important role on its biodegradation by MPO extracted from human neutrophils [33]. Comparing different GO samples, we found that MPO was able to completely degrade those that were highly dispersed.
In addition to the biodegradation approaches mentioned above, we have recently explored a novel strategy called 'degradation-by-design' of carbon nanomaterials [34]. This concept consisting on covalent functionalization with specific molecules, which can enhance the catalytic activity of peroxidases, has now been extended to graphene oxide. Two molecules, corresponding to 7-hydroxy azido coumarin (AZC) and 3,4-dihydroxybenzoic acid (DHBA) were covalently conjugated to the surface of GO. These specific ligands of HRP attached to GO are aimed to enhance the enzymatic action either by attracting the enzyme in close proximity of its active site or by mediating the electron transfer between GO and the enzyme, thereby leading to the increase of the oxidation rate. The degradation experiments using HRP have revealed that GO covalently conjugated with these two substrates has a better biodegradability over unmodified GO. In addition, we have also functionalized the surface of GO with triethyleneglycol diamine (TEG) to investigate the effect of free amine groups on the biodegradation process, which has resulted in slowing this process. Our results indicate that functionalization can modulate the degradation of GO, which can be enhanced by changing the functional groups on its surface. The design of GRMs functionalized with appropriate substrates will be fundamental in the conception of new and safer composites containing such materials.
Materials and methods
Synthesis of GO conjugates
Full details on the synthesis of the organic precursors and the different functionalized GO and their characterizations are reported in Supporting Information.
Enzymatic degradation of GO, GOAZC, GODHBA and GOTEG
GO, GOTEG, GOAZC and GODHBA powder (1 mg of each sample) were dispersed separately in 5 ml phosphate buffer saline (PBS) and sonicated for 2 min. HRP (1.2 mg) pre-solubilized in 2 ml of PBS was then added. To enable the enzymatic activity, 10 µl of a 0.1 m solution of H2O2 (final concentration 142 µM) were added once per day for all duration of the experiments. The control samples (without addition of HRP) of all GO conjugates were also prepared in the same way adding the same amount of H2O2 (final concentration 142 µM) every day for all duration of the experiments. All suspensions were stirred in the dark at r.t. for the entire duration of the experiments. Aliquots (200 µl) were withdrawn at 0, 7, 12 and 20 d and stored at −20 °C in the dark until characterization by different techniques. In the case of GO and GOTEG, the degradation experiments were carried out for 30 d, after refreshing HRP (1.0 mg) after 20 d.
Native polyacrylamide gel electrophoresis
The interaction of HRP with the surface of GO sheets was evaluated by native polyacrylamide gel electrophoresis (PAGE) after incubating HRP with the different conjugates. Briefly, 50 µg of each GO conjugate was dispersed in PBS (10 µl) and sonicated for 2 min. Then HRP (60 µg in 10 µl PBS) was added to each suspension and allowed to interact during 20 min incubation. At the end of the incubation time, Laemmli buffer was added to the mixture and the samples were loaded on a Mini PROTEAN®TGX™ 10% precasted gel (BioRad). Gel electrophoresis was then run under non-reducing conditions to evaluate the interaction between the enzyme and GO using a Mini-PROTEAN II apparatus (BioRad) and applying 150 V during 40 min. The gels were then fixed using aqueous solution (methanol 45% and 1% acetic acid) followed by staining with Coomassie Blue (2 h at room temperature) and scanning densitometry was performed with a GS-800 Calibrated Densitometer (BioRad). The densitometry analysis was performed using the ImageJ 1.48f software. The intensities of the bands corresponding to HRP after incubation with the different GO samples were expressed relatively to the band of HRP alone (i.e. considered as 100% of intensity).
Molecular modeling and docking of functionalized GO sheets to HRP
All GO models were generated using the building function provided by PyMOL (Script provided by the ERG research group and PyMOL Molecular Graphics System, Version 1.7.4 Schrödinger, LLC) [35]. Four different kinds of graphene oxide models were generated: GO, GOTEG, GOAZC and GODHBA. The functional groups (TEG, AZC, DHBA) were attached at the edge in the middle of the GO sheets. The horseradish peroxidase (HRP) structure was taken from the RCSB Protein Data Bank (PDBID: 1H5A, chain A).
The input files required for the docking (in pdbqt format) for both receptor and ligands were generated using the ADT tools package provided by AutoDock4.0. The docking was performed using AutoDock Vina [36]. We used the following parameters: a cubic box was built around the protein, with the center of the protein as center of the cube (x = 5.766, y = 5.952, z = 13.735 for HRP). The grid map was calculated automatically by the Vina software. The dimensions of the cube were 126 × 126 × 126 points as x, y and z sizes for both receptors, with a spacing of 0.4 Å between grid points. All other parameters were kept as default in AutoDock Vina. All docking experiments were launched on 4 CPUs. The docking results were analyzed using PyMOL for visual inspection. All conformations in each predicted binding sites were examined separately.
Results and discussion
Synthesis of graphene oxide covalent conjugates
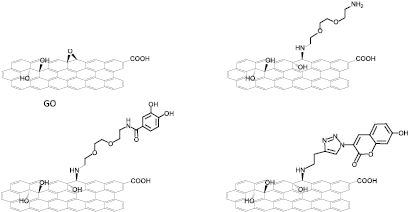
The starting GO used in this work was obtained from Grupo Antolin (Spain) in a powder form, easily dispersible in water. First, we prepared the triethylene glycol diamine conjugated GO (GOTEG) via epoxide ring opening reaction (figure 1) [37, 38]. This GO conjugate was used as control in the biodegradation study. Indeed, TEG linker has been introduced between the GO surface and catechol in GODHBA conjugate (see below). The amount of amino functions was measured by Kaiser test resulting in 191 µmol g−1 of GOTEG [39, 40]. FTIR spectroscopy analysis of GOTEG confirmed the covalent conjugation of TEG diamine to GO (figure S1) (stacks.iop.org/TDM/5/015020/mmedia). This was also supported by TGA (figure S4). Next, GO was treated with 3,4-dihydroxybenzoic acid (catechol moiety) linked to diaminotriethylene glycol (compound 5, DHBA-TEG-NH2; see Supporting Information for the preparation and characterization), leading to the formation of GODHBA (figure 1). The coupling of DHBA-TEG-NH2 to GO was confirmed by FTIR spectroscopy (figure S2) and TGA (figure S5). In the FTIR spectrum of GODHBA (figure S2), the absence of epoxide peak at 1230 cm−1 confirmed the ring opening of the epoxides by DHBA-TEG-NH2. In addition, peaks at 2914 and 2850 cm−1 correspond to C–H vibrations of the methylene groups of TEG linker.
Figure 1. Molecular structures of the different covalently functionalized GO conjugates. For simplicity, the structures show only one functional group in each GO sheet. The functional groups are reported in bold.
Download figure:
Standard image High-resolution imageTo explore the possibility of other HRP ligands, we prepared a second conjugate containing a coumarin derivative. Initially, GO-Alkyne was synthesized via epoxide ring opening by treating GO with 1-amino-3-butyne, similarly to GOTEG. GO-Alkyne was characterized by FTIR spectroscopy and TGA (figures S3 and S6). Then, we conjugated 3-azido-7-hydroxycoumarin (AZC) to GO-Alkyne by click chemistry (see experimental section for details) [41]. The strong peak at ~1605 cm−1 and the small peak at 1077 cm−1 in the FTIR spectrum of GOAZC (figure S3) are indicative of the triazole ring [42–44].
In order to characterize the dispersibility and the stability of the different samples, we measured the zeta potential (ξ) of all GO conjugates dispersed in water at pH 7. We found that GO suspension had ξ = −40.0 mV (standard deviation (SD): 1.1), which was significantly increased to −28.7 mV (SD: 3.05) in the case of GOTEG. This is due to the introduction of a certain amount of amino functions on the surface of GO. The presence of amino functions in GOTEG sample was confirmed by the positive surface potential at pH 2, ξ = 5.23 mV (SD: 1.4). The reduction of ξ is due to a charge compensation between the positively charged amino groups and the high concentration of oxygenated species still present on the surface of GOTEG. The zeta potential of GOAZC and GODHBA was −28.33 mV (SD: 1.36) and ξ = −33.13 mV (SD: 1.49), respectively. The slightly lower negative value for GODHBA could be due to the additional two hydroxy groups of DHBA. Next, the morphology of all GO conjugates was analyzed by TEM (figure S7). The flat 2D morphology was maintained after functionalization in the case of GOAZC and GODHBA (figures S7(B) and (C), respectively), whereas in the case of GOTEG moderately aggregated sheets were observed (figure S7(D)).
Biodegradation of GO, GOAZC, GODHBA and GOTEG
The main objective of this study was to assess the capacity of the functional groups on GO to enhance the biodegradability of the material. For this purpose, we carried out the degradation experiments with all GO conjugates by incubating with HRP followed by the addition of hydrogen peroxide once in a day to maintain the same catalytic activity along the entire period [24, 29]. Briefly, 1 mg of each sample was dispersed in 5 ml of PBS (figure S8) and treated with HRP and H2O2. The first sign of degradation was given by the change in the color of the GO suspensions. In particular, there was a significant reduction in the color intensity of GOAZC and GODHBA suspensions after 20 d (figure S8), whereas moderate color changes were instead observed for GO and GOTEG suspensions even after 20 d. We decided to continue further the degradation of GO and GOTEG up to 30 d. After this period these suspensions were still not translucent suggesting incomplete degradation. The disappearance of the dark brownish color of GOAZC and GODHBA was attributed to the degradation of GO conjugates by enzymatic catalysis [24, 29, 32]. These initial observations suggested that GO functionalized with catechol and coumarine derivatives was degraded faster than GO, while the addition of diaminotriethylene glycol likely slowed down the rate of degradation.
Size distribution analysis by dynamic light scattering
To confirm the visual observations, we applied a series of analytical, spectroscopic and microscopic techniques. The changes in the dimensions (i.e. lateral size) of all GO conjugates before and after treatment with HRP/H2O2 was followed by dynamic light scattering (DLS) [33]. The size of GO dispersed in PBS before the degradation was comprised between ~800 and 3000 nm (figure 2) with few traces of material around 10 µm. The dimensions were reduced to ~400–600 nm after 30 d treatment with HRP. These data suggest that the degradation of this type of GO by HRP was occurring only moderately. The size of GOAZC passed from ~1000 nm to ~300 nm, after 20 d. In the case of GODHBA, 20 d action of HRP resulted in a clear reduction of the sheets size mostly below 100 nm. A slight effect was instead evidenced on GOTEG sheets even after 30 d (variation from 700–1500 nm to 400–650 nm), indicating that the degradation has not taken place effectively. Overall, DLS studies supported that the degradation was effective and much faster in the case of GOAZC and GODHBA over GO and GOTEG, respectively. The degradability trend derived from DLS analysis is: GODHBA ⩾ GOAZC > GO > GOTEG, in good agreement with the color changes of the suspension. Simultaneously, to assess the effect of H2O2 alone on degradation, DLS measurements were also performed on all GO conjugates treated with hydrogen peroxide. In this case the dimensions of the samples were not affected (figure 2, last column) [33, 34].
Figure 2. DLS size distribution of GO, GOAZC, GODHBA and GOTEG suspensions. First and second columns show the size before and after the treatment with HRP/H2O2, respectively, at the different time points, while the third column displays the size of the control samples after treatment only with H2O2 for the same periods. (d. = diameter.)
Download figure:
Standard image High-resolution imageRaman spectroscopy analysis
Next, we employed Raman spectroscopy to obtain further insights about the effects of the degradation process on the structure of the GO conjugates during the enzymatic treatment. Raman spectroscopy was used for the systematic analysis of the oxidation level of graphitic materials during their degradation by various peroxidases [26, 31–33]. Raman spectra of all GO conjugates at the different time points of degradation were recorded (figure 3). The degradation state or oxidation level of GO was quantified by measuring the intensity ratios of D/G bands in each spectrum [45]. Figure 3(A) shows Raman spectra of GO. The characteristic D (~1350 cm−1) and G (~1600 cm−1) bands of GO are clearly visible with D/G ratio of 0.91 at day 0 [33, 46]. After 7 d treatment with HRP/H2O2, D/G ratio was increased to ~1.03 and reached ~1.21 after 12 d. After 20 d, the intensities of D and G bands were significantly reduced compared to the beginning, almost reaching the baseline by day 30, however maintaining a similar D/G ratio (1.02). In the same interval of time, we also observed a few distinct spectra showing high intense D and G bands (figures S9(A) and (B)), revealing that the degradation of GO was not uniformly occurring and incomplete even after 30 d. This result is not in agreement with the previous report by Star and co-workers [29], where after 20 d complete degradation of GO was observed by HRP. The dissimilar results can be ascribed to the type of GO used in the two studies [29, 33]. GO previously reported [29] was synthesized using well-known Hummers method, while GO used in this work was commercially produced from carbon fibers. Both GOs can vary in the amount of oxygen functional groups and aqueous dispersibility [33], which could explain their different degradability by HRP.
Figure 3. Raman spectra of GO (A), GOAZC (B), GOTEG (C) and GODHBA (D). Each line is an average of minimum five spectra.
Download figure:
Standard image High-resolution imageRaman spectra of GOAZC at day 0 had D/G ratio of 0.79 (figure 3(B)). After 7 d, a significant reduction in the intensities of D and G bands was observed, leading to an increase of D/G ratio to 0.84, sign of enzymatic degradation. Twelve day treatment resulted in the disappearance of both bands which was complete by 20 d. This result strongly evidenced how the degradation of GOAZC was much faster and complete compared to GO. In the case of GODHBA, D/G ratio was 0.99 at day 0 (figure 3(C)). After 7 d treatment, the D/G ratio reached 1.14, with a significant decrease and broadening of D and G bands. After 12 d, the intensities of both bands were negligible, revealing a complete degradation of this conjugate. This was confirmed at day 20. Finally, GOTEG had D/G ratio of 0.88 at day 0 (figure 3(D)). D/G ratio reached 1.06 after 7 d and slightly increased to 1.10 at day 12, while the intensities of D and G bands gradually decreased. Between day 20 and 30, two kinds of spectra were observed. In the first type, both D and G bands were less intense but D/G ratio was almost unchanged (~1.07). The second type corresponds to spectra with high intense D and G bands (figures S10(A) and (B)). This result indicates that the degradation of GOTEG is incomplete even after 30 d. The degradation results seem to be slower than starting GO.
In view of these results, we concluded that the specific functional groups on the surface of GO are playing a crucial role in the activity of the enzyme in the presence of hydrogen peroxide. During the treatment with HRP, the increase in the D/G ratio was dependent on the time of treatment, suggesting the formation of defects on the graphitic surface of respective GO sheets due to the oxidation caused by HRP catalysis [29]. The trend for the biodegradability of GO conjugates from the Raman spectroscopy analysis follows as: GOAZC ⩾ GODHBA > GO > GOTEG, consistent with DLS observations. We also assessed the D/G intensity ratio for all the control GO samples treated only with H2O2 without HRP. After 20 d, the D/G ratio was 1.05, 0.94 and 1.02 for GO, GOAZC and GODHBA, respectively, while it was 1.05 for GOTEG treated for 30 d (figure S11). In contrast to HRP treatment, the incubation of the GO conjugates with H2O2 alone for the same time did not affect significantly neither the D/G ratio nor the intensity of the D and G bands compared to the values at day 0.
Transmission electron microscopy analysis
To investigate the morphological changes during the degradation, we characterized all the GO conjugates at different time points by TEM. The starting GO sheets at day 0 with HRP are displayed in figure 4. After 12 d incubation, porous GO sheets started to appear, gradually increasing until day 20. However, the degradation of GO was not complete, since we still observed some aggregated sheets (figure S12). Additional 10 d treatment with HRP did not change significantly the morphology of the GO sheets (figure S13). In the case of GOAZC (figure 4), after 12 d HRP treatment, most of the sheets were converted into 'holey sheets' and broken down into tiny fragments. However, all these porous sheets were transformed into nanoparticles after 20 d. For GODHBA (figure 4), the degradation was more significant after 12 d, where complete rupturing of the sheets into tiny fragments was observed. By 20 d, all these tiny fragments were mostly absent revealing nearly complete degradation of GODHBA. In the case of GOTEG (figure 4), the size of the sheets was reduced only after 20 d, showing moderate porous morphology along with many aggregated sheets (figure S14), indicating that GOTEG was not degraded uniformly, similarly to GO. These results were also observed with Raman spectroscopy analysis of GOTEG. On other hand, the control experiments performed by only adding H2O2 to the respective GO samples did not affect sheet morphologies of any of the samples (figure S15).
Figure 4. TEM images of GO, GOAZC, GOTEG and GODHBA during the degradation by HRP/H2O2 at different time points. Scale bars correspond to 500 nm.
Download figure:
Standard image High-resolution imageOverall, the TEM results were in good agreement with DLS and Raman analyses, showing GOAZC and GODHBA much quicker and effective degradation than GO and GOTEG. In addition, the combination of Raman, DLS and TEM indicated the key role of the specific functional groups (AZC and DHBA) in the degradation of GO. Since both catechol and coumarin derivatives are known to be good reducing substrates for HRP, they clearly favor the interaction with the enzyme leading to the effective degradation of carbon materials, as we demonstrated for MWCNTs functionalized with the same molecules [34]. We can hypothesize that the presence of free amines on the surface of GOTEG reduces the interaction between GO and HRP due to electrostatic repulsion. To support this hypothesis, we studied the interaction between GO and HRP by gel electrophoresis and molecular modeling.
HRP interaction with functionalized GO conjugates
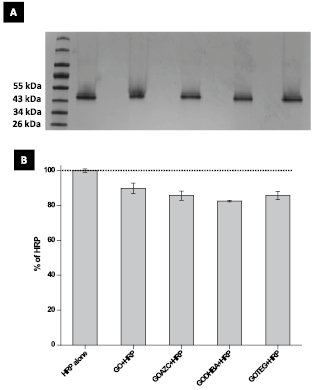
A native polyacrylamide gel electrophoresis (PAGE) was carried out to acquire more information on the interaction of HRP with GO conjugates. HRP was incubated with each GO conjugate for 20 min before running the gel. Some differences in the respective bands in the gel were observed for each GO conjugates (figure 5(A)). In fact, the densitometry analysis (figure 5(B)) shows that all GO conjugates are able to slightly reduce the migration of HRP, confirming the reduced intensity of HRP gel band at ≈44 kDa (figure 5(A)) [29]. This analysis suggests that functionalized GODHBA interacted with HRP a little better than pristine GO, likely because the additional hydroxyl groups of DHBA may increase the electrostatic interaction between the sheets and protein. Previous reports revealed that there is a specific interaction between GO [29, 47], or carboxylated CNTs [48, 49] and HRP compared to reduced GO or pristine CNTs. Our analysis suggests that HRP was adsorbed onto the GO sheets dependent on the surface functionalization, influencing, consequently, the enzymatic degradation.
Figure 5. (A) Gel electrophoresis of HRP alone or HRP incubated with the different samples of GO. Lane 0: protein ladder; lane 1: HRP alone (60 µg); lane 2: GO (50 µg) incubated with HRP (60 µg); lane 3: GOAZC (50 µg) incubated with HRP (60 µg); lane 4: GODHBA (50 µg) incubated with HRP (60 µg); lane 5: GOTEG (50 µg) incubated with HRP (60 µg). (B) The relative densitometry analysis of the HRP band (44 kDa): the results are expressed relatively to the band of HRP alone which is considered as 100% of intensity.
Download figure:
Standard image High-resolution imageMolecular modeling studies
Finally, we performed molecular docking studies to better understand the interaction of HRP with the different surface modified GO conjugates. The complexes between GO sheets and HRP are illustrated in figure 6. On the basis of the simulation, the closest distance between the heme active site of HRP and each GO conjugate sheet was measured. For the GO/HRP complex, we found a distance of 14.8 Å, which is slightly higher than the value reported previously for the same interaction (12.8 Å) [29]. This is due to the difference in the size of the respective GO sheets and the amount of oxygenated groups present on the surface of GO. In the case of GOAZC, the nearest distance between the AZC molecule and the heme is 12.6 Å. The distance between DHBA molecule of GODHBA and HRP active site is 17.2 Å. Although the distance is higher compared to GO/HRP, the GO sheet is nicely projected towards HRP. In the case of GOTEG, the position of the sheet is in a totally different side of the protein, far away from the active site of the enzyme (>28.3 Å) likely due to a nonspecific interaction.
Figure 6. Molecular models revealing the possible binding positions of HRP to functionalized GO samples. Specific functional groups are shown in bold.
Download figure:
Standard image High-resolution imageAs discussed earlier, the electrostatic repulsion between the positively charged amine groups of GOTEG and the cationic nature of HRP plays a major role to keep graphene far away from the heme site. This result was also supported by gel electrophoresis (figure 5). The molecular docking study revealed that HRP could bind more tightly to GOAZC over GO. Because of the projection of 7-hydroxy azidocoumarin group conjugated to GO, the shortest distance between the heme active site and the surface of GOAZC was attained (figure 6). In addition, we also identified the number of amino acid residues of HRP within 5 Å proximity to the surface of all GO conjugates (table S1), since these residues were found earlier to affect the degradation process [29]. Compared to GO, there are 9 additional amino acid residues for GOAZC and 5 extra residues around GODHBA. Overall, we believe that these additional residues could lead to a better binding of HRP to GODHBA and GOAZC compared to GO and GOTEG. The vicinity and the position of the different GO conjugates in respect to the enzyme, as assessed by the simulations, further confirm the degradability of the GO samples and the speed of the process.
GO degradation mechanism
The different characterization methodologies presented in this study confirmed that the degradation of GO functionalized with coumarin and catechol moieties was faster and almost complete in comparison to native GO and amine-functionalized GOTEG. Previous studies highlighted that GO or oxidized CNTs should be in close proximity to the heme active site of HRP for more effective degradation [24, 31]. Our molecular models revealed that GOAZC and GODHBA have additional amino acid residues of HRP within 5 Å distance compared to GO and GOTEG, which lead to a better interaction with HRP, as also supported by gel electrophoresis. Hence, the combination of close proximity to the heme-active site of HRP and additional amino acid stabilization could be responsible for a more effective degradation of GOAZC and GODHBA in comparison to GO and GOTEG. In addition, most of these amino acids participated in the catalysis of oxidation reactions [24, 50, 51]. To highlight the importance of the covalent functionalization to enhance the biodegradation of GO conjugates, we have performed a control experiment using a non-covalent mixture between GO and DHBA. The simple presence of DHBA in solution does not accelerate the degradation process of GO, proving that the ligand needs to be covalently attached to the surface of the nanomaterial (figure S16). Raman spectroscopy analysis showed the increase in D/G ratio as well as the broadening of D and G bands during HRP treatment. This is due to the gradual increase of defects on the surface of GO conjugates along with the cleavage of C–C or C–O bonds by radical intermediates of HRP [52–54]. On the other hand, oxidation of GOAZC and GODHBA could start via either oxidizing the functional molecules (coumarin or catechol) attached to GO or enhancing the enzymatic activity of HRP, since both were proven to be efficient reducing substrates for HRP [55–60]. The free OH group in the position 7 of benzopyran ring (AZC) seems to play an important role in the enhanced degrading action of HRP, likely acting as an effective donor of radical species [23, 34, 48, 61]. Catechol derivatives (DHBA) act as redox mediators by favoring electron transfer, thereby preventing enzyme inactivation [34, 57, 62]. Recently, we have also demonstrated that AZC and DHBA conjugation to MWCNTs had improved degradation over oxidized MWCNTs [34]. We can conclude that the degradation of GODHBA and GOAZC also followed a similar pathway of structural decomposition.
Conclusions
We have demonstrated that biodegradation of GO is strongly dependent on the type of functional molecules attached to its surface. GO functionalization with specific molecules can enhance its biodegradation due to a better interaction between GO and HRP (i.e. coumarin derivatives) or via better electron transfer between the enzyme and GO (i.e. catechol derivatives). The amino functions on GO, although they did not change a lot the zeta potential, have instead a negative effect on the biodegradation. Overall, DLS, electron microscopy and Raman spectroscopy analyses are in good agreement and all supported the enhanced degradation of GO functionalized by coumarin and catechol moietis. Molecular simulation studies have confirmed the role of the specific functional groups for a better binding of GO sheets to protein compared to unmodified GO. This study also suggest how specific functionalities can help to increase biodegradability of GO by other peroxidases, leading to the design of efficient biodegradable carriers based on graphene nanomaterials.
Acknowledgments
This work was supported by the Centre National de la Recherche Scientifique (CNRS), by the Agence Nationale de la Recherche (ANR) through the LabEx project Chemistry of Complex Systems (ANR-10-LABX-0026_CSC) and by the International Center for Frontier Research in Chemistry (icFRC). The authors gratefully acknowledge financial support from EU FP7-ICT-2013-FET-F GRAPHENE Flagship project (No. 604391). We wish to acknowledge Petra Hellwig and Frédéric Melin for giving access to Raman spectroscopy instrument, Cathy Royer and Valérie Demais for TEM analyses at the Plateforme Imagerie in vitro at the Center of Neurochemistry (Strasbourg, France). Grupo Antolin (Spain) is also greatly acknowledged for providing the material.