Abstract
MRI is often the most sensitive or appropriate technique for important measurements in clinical diagnosis and research, but lengthy acquisition times limit its use due to cost and considerations of patient comfort and compliance. Once an image field of view and resolution is chosen, the minimum scan acquisition time is normally fixed by the amount of raw data that must be acquired to meet the Nyquist criteria. Recently, there has been research interest in using the theory of compressed sensing (CS) in MR imaging to reduce scan acquisition times. The theory argues that if our target MR image is sparse, having signal information in only a small proportion of pixels (like an angiogram), or if the image can be mathematically transformed to be sparse then it is possible to use that sparsity to recover a high definition image from substantially less acquired data. This review starts by considering methods of k-space undersampling which have already been incorporated into routine clinical imaging (partial Fourier imaging and parallel imaging), and then explains the basis of using compressed sensing in MRI. The practical considerations of applying CS to MRI acquisitions are discussed, such as designing k-space undersampling schemes, optimizing adjustable parameters in reconstructions and exploiting the power of combined compressed sensing and parallel imaging (CS-PI). A selection of clinical applications that have used CS and CS-PI prospectively are considered. The review concludes by signposting other imaging acceleration techniques under present development before concluding with a consideration of the potential impact and obstacles to bringing compressed sensing into routine use in clinical MRI.
Export citation and abstract BibTeX RIS

Content from this work may be used under the terms of the Creative Commons Attribution 3.0 licence. Any further distribution of this work must maintain attribution to the author(s) and the title of the work, journal citation and DOI.
1. Introduction
MRI has revolutionized clinical medical imaging and transformed medical research by providing repeatable, non-invasive measurements of tissue structure and function. MRI is uniquely flexible as the sensitivity of the image to tissue properties can be extensively varied: this can be done by altering the timing with which MR signals are collected (the echo time, TE and repetition time, TR), or by the use of magnetization preparation or contrast agents. These techniques provide contrast in standard anatomical MRI, but can also allow measurement of function in both clinical and research settings in most of the tissues and organs of the human body. Such measurements have reduced the need for invasive techniques such as tissue biopsy or radionuclide studies. This non-invasiveness means that it is feasible to use quantitative MRI as a longitudinal biomarker in trials of therapy (Hollingsworth 2014, Macauley et al 2015). MR methods have come to represent gold standards of measurement for clinical research and yet the modality remains relatively unused. Why? A common refrain is that 'MRI is difficult and expensive'.
In MRI research studies, the critical factors are the cost per volunteer of the imaging (dictated principally by scan duration) and the ability of study subjects to comply with scan procedure: this is influenced by scan duration, since subjects have to lie still throughout an acquisition in order not to degrade the quality of the images. Additionally, subjects must hold their breath for abdominal/thoracic imaging, which may be difficult for children, obese individuals and those with respiratory compromise. In designing clinical study protocols, a balance must be achieved between the physiological parameters that could be measured, the demands made of the patient and the cost of the scan session. The cost of MRI can vary widely, depending on the nature of the staffing and infrastructure costs supported by the charges (Fletcher et al 1999, Lewis et al 2015). In the UK the cost of performing MRI research in a university teaching hospital is typically in the range of £350–£500 per hour of scanner occupation, depending on the staffing and infrastructure costs supported. Throughout the history of clinical MRI there have been progressive improvements in image quality and functional measurement together with reductions in scan duration, through ingenuity in hardware and software development.
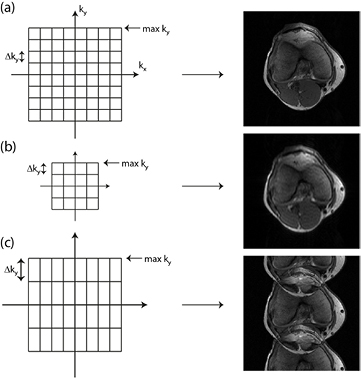
The image information in MRI is not acquired directly in the image space but rather in k-space, which contains information about spatial frequencies within the image. The image space and k-space are related by Fourier transformation. Once we have prescribed the field-of-view (FOV) and spatial resolution of the image that we wish to obtain, the k-space information that we must then conventionally acquire is determined by the Nyquist criterion (Nyquist 1928). The distance between neighbouring points of k-space is inversely proportional to the field of view in each direction (figure 1), while the highest frequency acquired in each direction is inversely proportional to the desired resolution. The k-space is filled by acquiring data which are encoded using pulsed magnetic field gradients. In one direction, known as the read direction (usually picked to be the longest direction of the anatomy, in figure 2 right–left), we can acquire a line of k-space points very rapidly within one repetition time using either a spin or gradient echo. However, further directions have to be phase encoded (in figure 2, the anterior–posterior and foot-head directions), which requires one repetition time to encode one line of k-space points. This has then to be repeated for every combination of the number of phase encoding steps required in the anterior–posterior and the foot-head directions. Therefore, MRI acquisitions can be lengthy, particularly those prescribed with high resolution or with wide coverage.
Figure 1. The relationship between k-space and image space for regular Cartesian sampling. (a) A fully sampled ky–kx space where max ky α 1/(y-resolution) and Δky α 1/(field of view in y direction), similarly in the x direction. (b) If we half the max ky and kx but preserve Δky and Δkx then we have halved the resolution of the resulting image while preserving its field of view. (c) By contrast, if we preserve max ky and miss out alternate phase encoding lines, thus doubling Δky, we preserve the resolution but half the field of view in the y direction leading to fold-over.
Download figure:
Standard image High-resolution imageFigure 2. Once a matrix size is chosen for a 3D acquisition, the Nyquist condition enforces the collection of data to fill an equivalently sized k-space matrix, where the maximum encode is related to the reciprocal of the resolution and the spacing to the reciprocal of the field of view. In the absence of any acceleration, each line in the phase encoding direction will require one repetition time (TR) to acquire. In this example, there is rapid frequency encoding right–left (256 encodes) and slower phase encoding in the anterior–posterior (190 encodes) and foot-head directions (48 encodes, of which 5 selected planes are shown). 190 × 48 phase encodes are required to acquire the signals for this prescription.
Download figure:
Standard image High-resolution imageThroughout the history of MRI there have been innovations which have tried to overcome the limitation of the repeated repetition times, including echo planar imaging (EPI) (Mansfield 1977), rapid acquisition with relaxation enhancement (RARE, also known as fast or turbo spin echo) (Hennig et al 1986), and fast low angle shot imaging and its variants (Haase et al 1986). Rather than the simple acquisition described above, where only one line of k-space is acquired from one gradient or spin echo per repetition time1, these techniques use multiple rf pulses or gradient refocussings to generate multiple echoes and acquire multiple lines of k-space per repetition time: a summary of the different techniques in common clinical use can be found in (McRobbie et al 2006, pp 220–56). These techniques accelerate the acquisition of the full k-space coverage demanded by the Nyquist criterion and can therefore still be regarded as accelerated fully sampled acquisitions.
An alternative or additional method of reducing the acquisition time is to not collect the full number of k-space points demanded by the Nyquist criterion. Such an acquisition is then said to be undersampled. In Cartesian imaging, as k-space points in the read direction are acquired very quickly, there is generally little advantage to undersampling in the read dimension. Rather we omit the collection of whole lines of k-space in one of the phase encoding directions. If a straightforward inverse Fourier transform is applied to such data then artefacts will be seen in the reconstructed image, the precise nature of the artefact depending on the undersampling pattern. However, if we can bring additional information to stabilize the reconstruction, then we may be able to produce images of acceptable quality from sub-Nyquist acquisitions. This basic premise underlies the techniques of partial Fourier imaging, parallel imaging and compressed sensing reconstruction. The first two of these methods are now routinely available on all clinical scanners and have been adopted by radiologists for routine diagnostic imaging.
In their present implementation on clinical scanners, the first two methods differ from compressed sensing in that they omit phase encoding steps in a regular fashion: compressed sensing demands an irregular undersampling pattern that will not lead to aliasing artefacts in the image domain, which we shall define further in section 3. The idea of irregular undersampling of data followed by a constrained reconstruction has been studied in general signal processing for some time (Donoho 2006), but the concept of using this within MRI was brought to the attention of the MR physics community by a number of papers including Lustig et al (2007) and Block et al (2007). Since then the number of papers with different undersampling and reconstruction schemes involving compressed sensing has burgeoned mightily, though as we shall see, only a few of these offer practical solutions for clinical scanning.
In this review, we will first examine today's most common methods of acceleration, partial Fourier imaging and parallel imaging (section 2), before going on to explain the principles of compressed sensing (section 3). We will then examine the practicalities of implementing compressed sensing (section 4), including the necessity of determining optimal reconstruction parameters, choosing the degree of acceleration to use and the computational burdens of the reconstructions. It is also critical to consider how we should validate MR data that have been reconstructed from undersampled data using compressed sensing, for both radiological and quantitative research applications. Applications which have been validated are then reviewed (section 5). There are alternatives to irregular undersampling and compressed sensing reconstruction which may be better suited to accelerate the acquisition of certain image types (section 6), and we conclude by considering the future challenges that remain to be overcome to gain widespread acceptance for acceleration by compressed sensing (section 7).
2. Acceleration by partial Fourier imaging and parallel imaging
2.1. Partial Fourier imaging
If we are principally interested in the magnitude information of an MR image, and we do not require high resolution phase information, as is often the case in diagnostic radiology, then we can exploit the property that the Fourier transformation of a purely real function has complex conjugate symmetry in k-space (Feinberg et al 1986). In theory this means that we only have to acquire half of the k-space in the phase encoding direction and we can reduce the number of TRs required twofold. In practice, in order to provide robust phase correction slightly more than half (commonly ~60%) of the phase encodes are acquired. This reduces the acquisition time, though there is a corresponding fall in SNR. It is also possible to apply this in the read direction to reduce the echo time, which may permit a slight reduction in repetition time per phase encode.
2.2. Parallel imaging
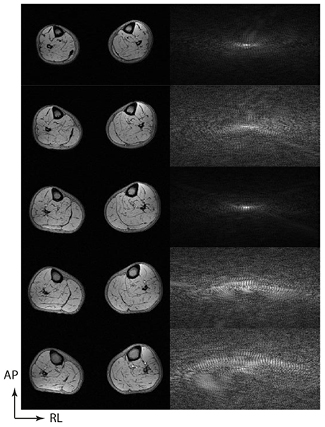
During the late 1990s the radiofrequency coils used to receive the signal in clinical MRI started to change in design, as it was realized that using an array of independent receiver channels could increase the overall signal to noise of the image compared to a single homogeneous volume coil (Roemer et al 1990). The other consequence of the phased array design is that each of the independent channels in the array is most sensitive to the tissue nearest to that coil (figure 3), and these sensitivity maps provide additional information which can be used to stabilize an undersampled image reconstruction.
Figure 3. Parallel imaging stabilizes sub-Nyquist acquisitions, which use a reduced field of view by using the sensitivity profiles from the individual elements of the phased array coil. Here a cross-sectional image through the lower legs is acquired using a 6-channel surface coil as shown (top). Beneath, the images derived from the signal from the 6 separate channels are shown, demonstrating maximal signal intensity nearest the coil element in question, with fall off of intensity elsewhere. This information can be used to stabilize the undersampled acquisitions.
Download figure:
Standard image High-resolution imageIf the k-space is undersampled in a regular manner by missing out every alternate line of k-space in a phase encoding direction, the number of repetition times required (and hence the total acquisition time) can be halved (figure 1(c)). The additional information about the coil channel sensitivities can be introduced by acquiring a separate low resolution scan, as is done in the sensitivity encoding (SENSE) technique (Pruessmann et al 1999) and calculating sensitivity maps which stabilize the reconstruction in image space, effectively by permitting the mathematical unfolding of aliased signal. The alternative is to fully acquire a small central portion of k-space, which corresponds to a low resolution image covering the same field-of-view as the main acquisition (figure 1(b)), and use the acquired points to calculate the missing k-space data before standard inverse Fourier transformation, as is performed in the generalized autocalibrating (GRAPPA) techniques (Griswold et al 2002).
Since the original introduction of these methods using regular undersampling with Cartesian acquisition, the techniques have been expanded to allow parallel imaging to be used with arbitrary sampling schemes and with non-Cartesian acquisitions (Pruessmann et al 2001, Seiberlich et al 2007, Lustig and Pauly 2010). In general, such techniques require iterative reconstructions and have not yet been widely used on clinical systems.
Parallel imaging methods have been found to be sufficiently robust for clinical imaging and at least one of the methods is now implemented on all clinical scanners, the choice of method being presently divided between vendors. The acceleration that can be achieved by these methods is limited by several factors. The first is that the arrangement of the coil elements has to provide varying sensitivity information in the same direction as the phase encodes to be omitted, and the acceleration factor cannot typically be greater than the number of elements in that direction (so a factor of 2 in the anterior–posterior direction for the coil in figure 3), subject to a factor related to the geometrical arrangement of the array, and thus how closely correlated are the coil sensitivities from different array elements. If this acceleration factor is exceeded, then coherent artefact will become apparent in the reconstructed image as well as enhanced noise amplification from the high acceleration factor. A detailed consideration of the possible artefacts is available elsewhere (Deshmane et al 2012). The second consideration, applicable to all k-space undersampling methods, is that a reduced acquisition of energy in k-space reduces the available signal to noise, and hence parallel imaging can only be used where there is sufficient signal to noise in the original fully sampled image.
3. Compressed sensing
3.1. Introduction
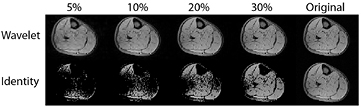
How then to explain how image acquisition can be accelerated by compressed sensing? First, let us imagine taking a picture with our digital camera. The sensors within the camera detect an image and signal intensity values are assigned to each of, say, 15 megapixels. However, storing the information for every pixel independently will lead to extremely large files, and so the camera instead mathematically converts the image to JPEG format (Taubman and Marcellin 2002). The JPEG 2000 format uses a mathematical transform known as a discrete wavelet transform to compress the information for storage at a smaller size. The discrete wavelet transform applies a nested series of low and high pass filters to the image, producing a wavelet space of the same matrix size as the original image space (Xiong and Ramchandran 2009). This property becomes most apparent if we examine the discrete wavelet transform of an MRI cross-section through the lower leg (figure 4(a)). Most of the pixels in the original image have non-zero signal intensity: the image is therefore not sparse. Some MR images are naturally sparse, such as angiograms, though the majority are not. It becomes sparse in the wavelet domain, since a smaller number of pixels (which are now wavelet coefficients) carry the important image content (figure 4(b)). The difference can be illustrated by ordering the image intensities and wavelet coefficients in order of absolute size (figure 4(c)). The image can be reconstructed with high fidelity from only the most significant coefficients in the wavelet domain, whereas this is not possible in the original image domain (figure 5). For real image objects, this is not a lossless compression, that is to say that the original data is not perfectly reconstructable from the reduced number of coefficients. Therefore, there will be a trade-off between the reduction in the number of wavelet coefficients used and the image fidelity required.
Figure 4. (a) Most MR images are not themselves sparse. In the image space, most of the pixels in the image contain important non-zero information. (b) Under the discrete wavelet transform, a lot of the important signal information has been compressed into a smaller number of pixels or wavelet coefficients with non-zero values, and there are many pixels with zero or near zero values. (c) If we put the absolute intensity values of the image intensity (green) and the wavelet coefficients (blue) in order we can see that the intensity of the wavelet coefficients tends towards zero much more rapidly. The wavelet representation is therefore sparse.
Download figure:
Standard image High-resolution imageFigure 5. Reconstructing images from a smaller number of coefficients. We take the intensity of the signal in the wavelet domain (top) or the image intensity (bottom) and retain only the highest intensity 5%, 10%, 20%, 30% of the signals. The remaining wavelets are retransformed to the image space. We can see that the wavelet space is much sparser than the original image space, with reconstruction from only 20% of the wavelet coefficients giving reasonable image definition compared to the original (right).
Download figure:
Standard image High-resolution imageSo how can we move from an issue of image storage to actually reducing the acquisition time? If we are able to describe an MR image in a transform space in which it is sparse, then the compressed sensing theory argues that we may be able to reconstruct that image from a smaller number of measurements in k-space, provided that the k-space undersampling is performed in such a way that the artefacts in the image domain do not give rise to structured aliasing. This is a direct contrast with the sampling patterns required for conventional parallel imaging (section 2.2 and figure 1(c)), which are regular undersampling patterns and give rise to structured aliasing of the signal under inverse Fourier transform.
Consider the transverse image of the lower leg muscles in figure 6(a), which is derived from fully-sampled data. If we transform the image with a discrete wavelet transform, the signal energy is compressed into a small number of wavelet coefficients with significant intensity. If we apply an irregular undersampling pattern, such as that shown in figure 6(b), then the inverse Fourier transform now shows evidence of noise in the image domain, but no structured aliasing, and the degree of degradation increases with the degree of undersampling (figures 6(c) and (d)). The image degradation arises as there is leakage of signal energy from each pixel into neighbouring pixels in the image domain. This artefact is also not structured in the wavelet domain, the most significant wavelet coefficients appear near-identical for fully sampled and undersampled images. But we already know from figure 5 that by keeping only the most significant coefficients we can reconstruct a high-quality image. By enforcing sparsity in the wavelet domain we limit the reconstruction of the image to the highest intensity wavelet coefficients which permit the reconstruction.
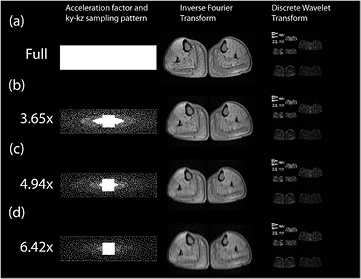
Figure 6. Examples of Cartesian variable density Poisson disk undersampling patterns with different accelerations. The phase encoding space of a 3D gradient echo (ky–kz plane) is shown for (a) a full Nyquist criterion acquisition and accelerations of (b) 3.65 ×, (c) 4.94 × and (d) 6.42 ×. In each case a Poisson disk with a quadratic fall off in sampling density has been designed, with a fully sampled centre portion to permit parallel imaging to be combined with compressed sensing using L1-ESPIRiT. In the middle column we see the effect of the sampling pattern if an inverse Fourier transform is performed. For (b), (c) and (d), there is no structured aliasing, but there is impaired image quality. Examining the discrete wavelet transforms of these acquisitions (right column) shows no discernible difference by eye.
Download figure:
Standard image High-resolution imageThis means that the sparsity of the original image in the discrete wavelet domain can be used to retrieve the original image from a small number of wavelet coefficients. This example enables us to see the pre-conditions of compressed sensing very clearly. If we had not used an irregular undersampling pattern, but had done a regular k-space undersampling such as is used in conventional parallel imaging, then the Fourier transformed image space would have contained structured aliasing, and this structured aliasing would have been contained, within the same highest intensity coefficients of the wavelet domain as the image proper, preventing us from recovering an artefact-free image by enforcing sparsity in the wavelet domain.
Our compressed sensing reconstruction must therefore satisfy two constraints. We need to make sure that the image that we are reconstructing is consistent with the undersampled k-space that we have acquired, and yet we need to enforce sparsity in the wavelet (or other) transform domain. Mathematically, this can be expressed as:


or equivalently putting these together using a Langrangian multiplier, λ,

where y is the measured k-space data, m is the reconstructed image, F is the Fourier transform, D is an operator which selected only those locations where undersampled raw data has been acquired, Ψ is the operator that applies the discrete wavelet transform to the image data while ε constrains how closely the reconstruction reflects the acquired data.
Critically, in the above equations, there are two types of norm being evaluated. The closeness of fit to the reconstructed data is being evaluated using the L2-norm  . This is a very familiar calculation to most quantitative scientists as it involves taking a 'least squares approach' to minimize the difference between the acquired data and the reconstructed image. Importantly, equation (1), which aims to maximize the sparsity of the image, uses not an L2-norm but an L1-norm,
. This is a very familiar calculation to most quantitative scientists as it involves taking a 'least squares approach' to minimize the difference between the acquired data and the reconstructed image. Importantly, equation (1), which aims to maximize the sparsity of the image, uses not an L2-norm but an L1-norm,  =
=  . In other words, it represents the absolute sum of all the elements within x. Strictly speaking we should penalize the solution based on the L0-norm, which is simply the total number of non-zero elements of x and hence a direct representation of the sparsity of x. However, minimising the L0 norm is a computationally expensive and unstable problem. Minimizing the L1 norm in equation (1) can be shown to enforce sparsity in x, reducing the number of non-zero components, while providing a tractable optimization problem (Baraniuk 2007). This is exactly what we need to retrieve the most significant non-zero coefficients from the wavelet space.
. In other words, it represents the absolute sum of all the elements within x. Strictly speaking we should penalize the solution based on the L0-norm, which is simply the total number of non-zero elements of x and hence a direct representation of the sparsity of x. However, minimising the L0 norm is a computationally expensive and unstable problem. Minimizing the L1 norm in equation (1) can be shown to enforce sparsity in x, reducing the number of non-zero components, while providing a tractable optimization problem (Baraniuk 2007). This is exactly what we need to retrieve the most significant non-zero coefficients from the wavelet space.
The nature of equation (3) means that we will need to perform an iterative solution method rather than a straightforward Fourier transform or a linear solution to a parallel imaging reconstruction. Numerous methods have been explored for solving these types of reconstruction problem, in particular to try and increase the efficiency of the minimization of the L1-norm. Coded examples of solutions by a non-linear conjugate gradient algorithm and an iterative soft thresholding algorithm have been made available by the authors of (Lustig et al 2007), which allow users to reproduce some of the reconstructions performed in that paper (at www.eecs.berkeley.edu/~mlustig/Software.html).
3.2. The sparsifying transform
In this discussion, we have considered the discrete wavelet transform as an example of a sparsifying transform, but it is only one of a great array of possible transforms, which can be used alone or in combination. Common choices that have been deployed include total variation, which is a summation of the gradients in intensity between adjacent pixels (termed 'finite differences'), discrete cosine transforms (which were used in the original JPEG standard) and many others. A pictorial comparison of cosine transforms, wavelet transforms and finite differences is shown in (Lustig et al 2007). Total variation has proved to be a powerful constraint either when used on its own (Block et al 2007), or when used as an additional constraint to a discrete wavelet transform (Wiens et al 2014). There is also a vast array of discrete wavelet transforms, of which the Daubechies-4 wavelet is commonly used, but many others have been used and assessed.
Adding a total variation constraint to a discrete wavelet transform can avoid artefacts from using wavelets alone (such as sharp transition bands within the image) due to the Daubechies wavelet not being translation invariant. However, the use of an additional total variation constraint can be avoided by performing randomized shifting of the wavelet transform with respect to the image (Figueiredo and Nowak 2003, Murphy et al 2012).
There is nothing to prevent other prior knowledge being incorporated through the addition of further constraint terms to the sparsifying transform. In a compressed sensing application to measure dynamic lung volume with high temporal resolution throughout the respiratory cycle, additional priors were added to impose knowledge of the image intensity distribution at end-inspiration and end-expiration (Berman et al 2015). However, the more terms of constraint that are added, the more complex the determination of the optimal values of the weighting multipliers becomes.
3.3. Combining compressed sensing reconstruction with parallel imaging and/or partial Fourier imaging
Several research groups have concentrated on synergistically combining the undersampling possible using compressed sensing and parallel imaging to achieve greater acceleration factors than by either technique alone, including (Block et al 2007, Liu et al 2009, Lustig and Pauly 2010, Murphy et al 2012, Uecker et al 2014). The problem to be solved can be expressed by modifying equation (3) to incorporate the sensitivity profiles, S, of the N receiver coils in the phased array. This then yields,

Two reconstruction methods for parallel imaging with arbitrary undersampling were proposed by the group of Lustig, SPIRiT (Lustig and Pauly 2010) and e-SPIRiT (Uecker et al 2014), which operate in the k-space and image domains, respectively, and can incorporate an L1-norm minimization term to additionally enforce sparsity in a transform domain. In order to derive the sensitivity map information using these methods, the undersampled k-space acquisition will need to either possess a fully sampled area at the centre of k-space, or for such data to be available from a previous acquisition. In all that follows, it will be assumed that the fully sampled centre portion of k-space is being acquired together with an undersampled peripheral k-space (figure 6), as this has been the most common way that combined compressed sensing and parallel imaging (CS-PI) has been performed.
As outlined in section 2.1, if only magnitude data are desired then partial Fourier encoding can be combined with both compressed sensing and parallel imaging (Liu et al 2012).
3.4. The design of undersampling schemes for compressed sensing reconstruction
How can we design the k-space undersampling schemes that we require for a compressed sensing reconstruction? We already know from section 2 that regular undersampling patterns will lead to structured artefact under inverse Fourier transform and are therefore unsuitable. The most desirable undersampling therefore will be quasi-random and will lack regular structure in the distance between the acquired k-space points and yet will not allow large regions where there are no acquired samples. In Cartesian sampling, the Poisson disk distribution achieves this by combining the random sampling of k-space while imposing constraints on the maximum and minimum distance between k-space points (Cook 1986). For signal-to-noise efficiency, the sampling pattern should also reflect the underlying signal density of k-space, which is not uniform but is most concentrated for the low-frequency components at the centre of k-space and falls away towards the periphery. Most practical Cartesian implementations of compressed sensing use a variable density Poisson disk to reflect this. A detailed algorithmic description of how such a variable density sampling pattern can be constructed for MRI can be found in (Gdaniec et al 2014).
Radial sampling has a particular appeal for compressed sensing as the spokes of a radial acquisition naturally lead to a dense sampling at the centre of k-space with decreased density towards the periphery. Undersampled radial acquisitions produce non-aliased blurring artefacts in the image space (figure 7) and therefore lend themselves well to compressed sensing reconstructions with a simple total variation constraint (Block et al 2007). This can be combined with golden ratio profile ordering (Winkelmann et al 2007), where the angle of the next radial line to be sampled is set to be 111.25° (which is 180° divided by the golden ratio 1.618) compared to the previous acquisition. This ensures that any consecutive selection of radial profiles that are retrospectively selected for reconstruction have the most favourable distribution, and that each new acquisition divides the maximum angle in k-space not covered by the previous profiles. Such properties not only permit robustness to anatomical motion, but provide an excellent basis for using combined compressed sensing and parallel imaging to reconstruct dynamic series of data (such as DCE-MRI) with a spatial and temporal resolution that can be selected post-acquisition according to the number of consecutive k-space spokes used per image (Chandarana et al 2013).
Figure 7. Radial undersampling produces non-aliased artefacts in the image domain. From left to right, we have the same image with 100% of the radial spokes required for the Nyquist limit (π/2 × matrix size), 20%, 10% and 5% of the required spokes. Image quality can remain good to low sampling levels with gradual development of non-aliased artefact. Good image quality can often be recovered using total variation constraints in reconstruction.
Download figure:
Standard image High-resolution imageSpiral undersampling also possesses similar non-aliased artefact and the benefit of collecting multiple data near the k-space centre for motion correction. In clinical imaging it has been applied in such applications as accelerated 4D flow imaging with CS-PI reconstruction (Dyvorne et al 2015) and to alter the sensitivity of variable density spiral fMRI (Holland et al 2013). The issue of more general design strategies for optimal Cartesian and spiral undersampled k-space trajectories has also been addressed (Seeger et al 2010) using Bayesian methods.
3.5. Spatiotemporal sparsity
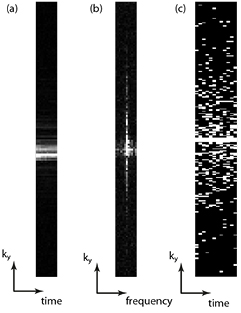
So far the discussion has concentrated on sparsity within static images. Clearly those image modalities where repeated images are taken with common image information, such as following the uptake in a DCE-MRI experiment using T1-w imaging, or measuring cardiac function by cine acquisition throughout the heartbeat, may have the potential for sparsity between the dynamics of the study. This means that additional constraints can be added to exploit this sparsity and that sparse sampling schemes can be spatiotemporal. If we take a slice of cardiac k-space data such that we see the ky phase encode versus time, then applying a Fourier transform in both the spatial and temporal dimension gives us a compact representation in an image–frequency (usually called y–f ) space (figure 8). If we then use a spatiotemporal sparse sampling pattern, which amounts to sampling different lines of ky in different temporal phases, then we can arrange to produce non-aliased artefact in the y–f space on which a sparsity constraint (such as minimization of the L1-norm) can be imposed to recover an artefact-free image. Examples of algorithms exploiting spatiotemporal sparsity include k–t FOCUSS (Jung et al 2007) and k–t SPARSE SENSE (Otazo et al 2010). Methods exploiting properties of compressed sensing are only a small subset of the temporal acceleration techniques available (Tsao and Kozerke 2012): in particular using a temporal Fourier transform is not necessarily the sparsest representation of the time domain and principal component analysis in the temporal dimension has also been implemented (Pedersen et al 2009).
Figure 8. (a) An intensity plot of the ky dimension versus time for a cardiac cine acquisition showing the similarity of the central k-space through the cardiac cycle. (b) If the time dimension is Fourier transformed, it can be seen that the resulting ky–f space is sparse and this can be used to constrain (c) a spatiotemporal undersampling scheme, here shown a with 4× acceleration factor.
Download figure:
Standard image High-resolution image3.6. Dictionary learning
The maximum acceleration factors that can be achieved is determined by the sparsity of the images in the transformed domain, and the discrete wavelet (and other fixed basis) transforms have been successful at sparsifying many different image types. However, additional sparsity might be obtained by the use of dictionary learning (Duarte-Carvajalino and Sapiro 2009), where the basis functions are learned from the data. A greater degree of sparsity would permit greater acceleration for a given reconstruction quality and this has become an important topic of recent research (Caballero et al 2014). The reconstruction burden for these methods is presently large and their general applicability and efficiency in MRI has yet to be proved.
4. Practical considerations for compressed sensing
There are several important practical considerations that must be taken into account to incorporate compressed sensing to our acquisition.
4.1. Adjustable parameters in CS reconstructions
A critically important consideration is that the reconstruction formulations detailed in section 3 have adjustable regularization parameters within them, which tune the trade-off between fidelity to the undersampled raw data and sparsity in the transform domain. The avid literature reader will find that many, if not most, researchers have performed an empirical search of the parameter space and 'appropriate' values of the regularization parameter are simply quoted, or perhaps the issue is simply omitted! In certain clinical applications optimal regularization parameters have been demonstrated that by comparing metrics of image quality between a fully-sampled acquisition and a sparsely sampled acquisition: such metrics include minimizing the root mean square error (RMSE) between the images or maximizing the structural similarity index (SSIM) (Loughran et al 2015, Mann et al 2015). It is found that for a given matrix size and protocol, the optimal reconstruction parameters are stable, and can therefore be confidently predicted for new scans using that exact protocol. There have been attempts to try and automate the choice of regularization parameter in some circumstances either through the adaptation of iterative soft thresholding algorithms (Khare et al 2012) or by more generalized reconstruction schemes (Aelterman et al 2014), which claim to require no adjustable parameters. These schemes, although potentially promising, have not yet been widely adopted or tested in clinical situations. The issue of successfully predicting appropriate reconstruction parameters remains a key limitation in permitting the use of compressed sensing in the general clinical environment.
4.2. Choosing the degree of acceleration to apply and validating CS reconstructions
What is the maximum degree of acceleration that can be applied in a given situation? There is no easy answer to this question, which will depend on the signal to noise of the base acquisition sequence, the imaging matrix size and the number of receiver coils used. The signal to noise matters since each accelerated acquisition only collects a fraction of the k-space energy of the original, the imaging matrix size is important as larger matrices provide greater scope to produce irregular k-space sampling patterns. The number of receiver coils has an influence as it determines the extent to which parallel imaging can be combined with compressed sensing to increase the overall acceleration.
Critically the purpose to which the images with compressed sensing images are put will determine the maximum acceleration. In the case of the reconstructed CS-PI muscle images shown in figure 9, the images appear qualitatively equivalent, though with increasing acceleration, there is some loss of high frequency features (such as minor fasciae), which make the muscle groups harder to differentiate. The data were acquired for the purpose of monitoring fat content in particular muscle groups and two independent observers concurred that a 5× acceleration was the maximum compatible with defining all the desired muscle groups (Hollingsworth et al 2014). However, had only the total muscle volume been desired then the 6.42× acceleration acquired (and possibly higher accelerations) would also have been acceptable.
Figure 9. Examples of combined compressed sensing and parallel imaging reconstructions of fat fraction maps for the calf muscle of a subject with Becker muscular dystrophy. The figure compares full Nyquist sampling with accelerations performed using variable density Poisson disk acquisitions at 3.65×, 4.94× and 6.42×. The reconstruction quality is found to be very good: in the broader series, some muscle groups cannot be segmented on the 6.42× acceleration images (Hollingsworth et al 2014).
Download figure:
Standard image High-resolution imageWhere the endpoints to be extracted are quantitative, it is possible to construct Bland–Altman analyses to compare the measurements from the accelerated images with those from conventional acquisition (Vincenti et al 2014, Loughran et al 2015, Mann et al 2015). According to the precision with which we wish to make the measurement and the segmentation required of the image, we can define the maximum acceleration.
Alternatively, if we are mainly concerned with visual fidelity, metrics such as the structural similarity index (Wang et al 2004) can be evaluated for increasing acceleration if we have both the fully sampled and the accelerated images (Hollingsworth et al 2014), or the parameters of a blurring filter (such as the full width half maximum of a Gaussian blurring kernel) (Ajraoui et al 2013). It may then be possible to define a lower bound of acceptability using such a metric. Much work remains to be done to generalize these findings.
Finally, for diagnostic imaging, the key test remains the reading of the subtle imaging feature of interest by two independent radiologists. The robustness of the technique to artefacts, the radiologist's understanding of those artefacts that do occur and determining that subtle yet important imaging features are not lost are critical to widespread acceptance.
4.3. Computational burden
It is important to recognize that image reconstruction using one of the compressed sensing formulations in section 3 requires the solution of an optimization problem involving the minimization of both L2 and L1-norms (equation (3)). This is in contrast to the simple inverse Fourier transform or the linear algebra required for a SENSE reconstruction. The solution may be computationally expensive, particularly if compressed sensing is combined with parallel imaging from phased arrays with large numbers of coils and/or multiple sparsity constraints are applied. The use of multi-core CPUs and fast GPU reconstructions can help to speed up the reconstruction times of Cartesian 3D CS-PI MRI using discrete wavelet transforms to a few seconds on relatively inexpensive computer hardware (Murphy et al 2012). In the context of the potential savings to be made in MRI scan time, an investment of £2000 in the computer hardware required for reconstruction is modest. For techniques such as radial CS-PI with motion compensation, the computational requirements can be extensive presently requiring 20 min or more and/or reconstruction on dedicated servers (Feng et al 2015). Traditionally, MR raw data have been reconstructed within the institution collecting the raw data and the transmission of data to remote powerful servers for immediate reconstruction may be hindered by both legal and technical considerations.
As mentioned above many phased arrays on modern systems may collect up to 32 channels of data. Immediate coil compression to a smaller number of virtual coils may help to reduce the computational cost of the iterative reconstructions required. There are many methods of doing this (such as Buehrer et al 2007, Doneva et al 2008) but one which exploits the fully sampled read direction is set out in (Zhang et al 2013), with accompanying code available at www.eecs.berkeley.edu/~mlustig/Software.html. The Lustig group has made a direct comparison of CS-PI reconstruction performed directly on data collected on a 32-channel coil and that with coil compression to 6 virtual coils, with no deterioration in image quality (Zhang et al 2014).
4.4. Availability of acquisition pulse sequences
To actually gain the benefit from the reduction in phase encodes and acquisition time, we will need to acquire only those phase encodes. Such irregular undersampling patterns, whether implemented as Cartesian, radial or spiral readouts are not commonly available on clinical scanners at the present time, and as with any novel technique within MRI practical implementation requires pulse programming access. There are now a number of research groups who have implemented pulse programmes which can perform prospective undersampling on clinical platforms, and much of this work is reviewed in section 5. This is presently a significant barrier to the development of CS, restricting it for the time being to dedicated MRI research groups, though advances in addressing the issues of reconstruction parameters and reconstruction burden may stimulate MRI scanner vendors to make these acquisition facilities available to their broader research base in the near future.
4.5. Resources for reconstruction code
A number of the authors of manuscripts on compressed sensing have made code available to allow others to reproduce the results of their research, and to permit users to explore the algorithms and the effect of varying parameter settings on their own data. Lustig's group has been notable for espousing this idea of reproducible research: that a paper describing a reconstruction algorithm is an advertisement for the scholarship rather than the scholarship itself. To this end, code to produce the results shown in several of the papers reviewed here is available at www.eecs.berkeley.edu/~mlustig/Software.html. Similarly, the reconstruction algorithms concerning radial acquisitions by the NYU group are available at http://cai2r.net/resources/software.
5. Prospective applications of compressed sensing
At the time of writing, a Pubmed search on the terms 'Compressed sensing AND MRI' yields over 350 manuscripts, which consider both the theory and practice of compressed sensing for many different MR contrasts, and in all parts of the body. Many of these manuscripts have concerned themselves with the refinement and comparison of different algorithms for performing the reconstruction from undersampled data, often by taking fully sampled data, retrospectively undersampling it and performing comparative reconstructions. In this section, in order to provide a perspective on maturing clinical applications using compressed sensing or combined CS-PI, we concentrate mainly on a selective review of those smaller number of publications where compressed sensing has been prospectively applied and evaluated, organized by MRI modality. The selection has been mostly restricted to clinical rather than preclinical studies.
5.1. General anatomical imaging
Perhaps because of the reasons outlined in section 4.2, where we consider the effects in image quality and appearance that can be caused by undersampling and reconstruction by compressed sensing, there has been more reluctance to trial its use for anatomical diagnostic imaging, with relatively few research papers on this theme. There is of course the selection bias inherent in that such methods are mainly available to research groups who tend to be interested in quantitative methods, rather than practising radiologists. Collaboration between the reconstruction group of Lustig and the clinical group of Vasanawala has led to the evaluation of combined compressed sensing and parallel imaging in pediatric imaging situations with independent scoring by radiologists in abdominal (Cheng et al 2014) cardiovascular and knee examinations (Vasanawala et al 2010). These sources are a good place to observe the practical benefits of CS-PI in one of the most challenging settings for MRI. Otherwise, there has been little applied work on standard clinical sequences, with one study applying compressed sensing to a 2D multi-slice brain protocol with modest acceleration factors (Sharma et al 2013), but with an acquisition which was probably not optimal for CS. Further clinical studies are awaited.
5.2. Fat fraction quantification and fat suppression
The use of MRI methods to quantify tissue fat fraction has increased substantially in recent years (Hu and Kan 2013). Methods such as the 3-point Dixon and IDEAL (iterative decomposition of asymmetric echoes with least squares estimation) to produce maps of proton density fat fractions which are independent of confounding factors such as magnet Bo inhomogeneity and radiofrequency coil inhomogeneity have found use in a number of applications. The most common is in the quantification of liver steatosis in NAFLD and type 2 diabetes, where breath-held measurements provide a means of longitudinally tracking the effect of interventions designed to ameliorate steatosis, whether low calorie diet, exercise regimes or pharmaceutical treatments. The quality of the fat fraction measurements hinges on fitting a model to the complex signal of a multi-echo gradient echo, which relies on the patient breath holding effectively for a duration of 17–30s.
Recently it has been demonstrated that combined compressed sensing and parallel imaging can be applied to 3D whole liver fat fraction measurements to reduce the breath hold from 17.7 to 4.7 s without substantial loss of image quality (figure 10): the high fidelity of the liver fat fractions extracted from the accelerated imaging was demonstrated by Bland–Altman analysis (Mann et al 2015). The same study also showed that the choice of weighting parameter required for the constrained reconstruction was consistent between all patients, which potentially permits reconstruction of sparsely sampled data without end-user interaction.
Figure 10. Hepatic fat fraction maps reconstructed from a 5-echo 3D IDEAL sequence using combined compressed sensing and parallel imaging. From the left to right we have a full acquisition, and undersampled acquisitions of 2.6 ×, 2.9 × and 3.8 ×, which lead to progressively shorter breath holds, with the ky–kz undersampling scheme given above the images. The hepatic fat fractions calculated from the volumetric scans are indicated beneath the images.
Download figure:
Standard image High-resolution imageHowever, where fat–water separation rather than fat fraction quantification is desired, simpler versions of these techniques, often using as few as two echoes, can be used to provide separation of the fat and water components of an image, providing more robust fat suppression than by selective B1 excitation, particularly at 3.0 T. (Gdaniec et al 2014) used combined CS-PI to perform water–fat separation in the abdomen. Rather than specifying the length of breath-hold, a CS-PI acquisition scheme was used in combination with a navigator that detected when the patient stopped holding their breath. The acquisition order was built up by using a centrally sampled area with concentric Poisson discs such that it was always possible to reconstruct an artefact-free image regardless of the breath holding capability of the patient, with those capable of longer breath holds providing higher resolution data.
Another important application of fat fraction measurement is the longitudinal tracking of disease progression in muscular dystrophies, where there is progressive replacement of healthy muscle with lipid (Hollingsworth 2014). Although there are no established cures for these genetic conditions, a number of therapeutic options have been developed, particularly for Duchenne muscular dystrophy, which require assessment over the short time scales feasible for first-in-man clinical trials. MR fat fraction measurement has been shown to be more sensitive than standard clinical measurements in demonstrating progression over short time periods (Willis et al 2013), and operating procedures for multi-national trials have been developed (Hollingsworth et al 2012).
The MR imaging forms but one part of very intense trials, where there is interest in a wide coverage of skeletal muscle, and often also of the heart, meaning that the burden of the MRI to often very young (5–10 year old) patients is substantial. Combined compressed sensing and parallel imaging MRI has been applied in subjects with Becker muscular dystrophy to achieve scan accelerations up to 5× over conventional fat fraction measurements (figure 9) to achieve full volumetric muscle coverage within a 1 min scan (Loughran et al 2015). Importantly it has been demonstrated that the adjustable parameters of the compressed sensing reconstruction are identical for a given undersampling pattern and acceleration, which means that these parameter values can be confidently used in the absence of fully sampled data. The image quality produced with a fivefold acceleration allows two observers to confidently delineate the muscle regions of interest used for analysis (Hollingsworth et al 2014).
5.3. Cardiac functional imaging
The mainstay of cardiac imaging is the balanced steady-state free precession cine sequence, retrospectively gated by VCG (vector cardiogram) R-wave monitoring, which is typically collected in 2D slices with view-sharing to reconstruct 25 phases throughout the cardiac cycle. This allows measurement of left ventricular mass, end-diastolic function and end-systolic function at reasonable spatial (1.3 mm × 1.3 mm × 7 mm) and temporal resolution (~40 ms depending on the heart rate). Typically this is acquired as a stack of say, 14 short axis slices, with 4 chamber and 2 chamber views, requiring as many as nine breath holds of ten seconds duration to collect the data plus additional breath holds to prescribe the anatomy. The recovery and preparation for these breath holds slows down the acquisition considerably, and is difficult for patients with respiratory compromise. Additionally, T1w sequences may be run with contrast agents to measure myocardial perfusion.
Many approaches other than compressed sensing have been taken to accelerate cardiac imaging and a comprehensive review of the field has been published elsewhere (Tsao and Kozerke 2012). Two more recent approaches combining compressed sensing and parallel imaging are of note. Firstly, there is the approach taken in Vincenti et al (2014), which shortens the acquisition by using a combination of CS-PI and partial Fourier encoding, but also reduces the number of cardiac slices to be acquired to 7 (4 non-contiguous short axis sections spread over the left ventricle and 3 long axis sections) by using an imposed guide-point model (Young et al 2000). This gave acceptable estimates of the main derived parameters while permitting the whole left ventricle functional parameters to be acquired in one 14 s breath hold.
Another recent approach taken to speeding up the standard cardiac examination is to combine CS-PI with motion correction to permit acquisitions to occur under free-breathing in a reasonable time frame. Radial acquisitions where every k-space trajectory passes through the origin of k-space lend themselves well to such schemes since they can be rebinned flexibly into different temporal, cardiac and respiratory states to optimize the reconstruction. Usman et al (2015) have recently demonstrated a whole left ventricular functional assessment from a 2 min free-breathing acquisition, taking advantage of a combination of CS-PI and motion correction, and demonstrating promising quantitative results on a small group of volunteers and patients. This is likely to prove particularly useful as the motion correction allows all the acquired data to be used in the reconstruction, in contrast to a similar Cartesian method (Xue et al 2013), which uses a respiratory navigator to reject a proportion of acquired data. Free-breathing coronary imaging with 100% scan efficiency has also been recently demonstrated (Prieto et al 2015). Due to the compatibility of free-breathing methods with compromized and uncooperative subjects, these approaches have the potential to find a place in routine cardiology.
5.4. Cardiac 4D flow imaging
The use of phase contrast to measure flow-rates in vessels has been used since the earliest days of clinical MRI. Initially this was performed by imaging a 2D slice perpendicular to the direction of flow and using a 1D phase encoding of flow to read out the velocity of the major vessels. However, temporally resolved volumetric measurement of flow patterns within the heart chambers and great vessels can be built up if phase encoding is applied in at least three orthogonal directions in combination with retrospective VCG gating and respiratory navigators, albeit at modest spatial resolution. Such patterns can be of great importance, particularly in understanding the alterations in aortic flow in artheroschlerotic disease and in the disruption of vortical flows within the heart chambers caused by congenital malformations (Parikh et al 2015). The preservation or dissipation of kinetic energy can be calculated. A recent review has surveyed the field (Stankovic et al 2014). However, acquiring the data for 3D + time flow information conventionally requires acquisitions which can be 30–40 min long. Such durations limit the applicability of these techniques and are unacceptable to patients if other MR modalities such as a high-resolution anatomical scan are also to be collected.
Combined CS-PI acceleration of 4D flow has been used prospectively in a number of applications (Hsiao et al 2012, Tariq et al 2013, Dyvorne et al 2014, 2015). The length of the full 4D acquisition means that it is difficult to perform global comparisons of the conventional and prospectively accelerated sequences, so these applications differ from the others considered in that there is usually no attempt to compare prospectively accelerated sequences with fully sampled sequences, but to use other available information. For example, the accuracy and precision of arterial and venous flow quantification by accelerated 4D flow MRI (mean duration 11 min) was assessed by examining conservation of flow between the superior and inferior venae cavae, the ascending and descending aorta and the pulmonary trunk (Tariq et al 2013). In another study, valvular insufficiency and shunts in young patients read from accelerated 4D flow MRI (mean duration 10 min) were compared with conventional cardiac MRI and echocardiography (Hsiao et al 2012).
Accelerated 4D flow MRI has also found application further from the heart, being used to assess portal vein and hepatic artery flow quantification (Dyvorne et al 2015). As in the cardiac applications, a CS-PI accelerated sequence with high spatial resolution was compared against a lower resolution acquisition with conventional parallel imaging. Whereas most of the techniques above have used Cartesian undersampling, a recent study has compared the use of undersampled spiral 4D flow measurements with Cartesian and found them to be better (Dyvorne et al 2014).
5.5. Dynamic contrast enhanced (DCE) imaging
DCE-MRI (including the study of cardiac perfusion) is an obvious target for compressed sensing techniques, since typically the same anatomy is imaged repeatedly with a T1-weighted acquisition sequence to monitor the change in T1 as a contrast agent enters the extracellular space of a given tissue or tumour from the vasculature. Using conventional fully-sampled imaging, there is a continual tension between the increasing the resolution or coverage of each individual imaging block to resolve fine anatomical detail and the need for fast acquisition to be able to accurately model the wash-in and wash-out of the contrast agent. These requirements become even more constrained if the organ of interest is affected by breathing motion (liver, kidney), since it is rarely possible to obtain the required data within a breath hold.
The study of (Chandarana et al 2013) is an example of the power that can be exploited by the use of undersampling. By using combined compressed sensing and parallel imaging on a radial acquisition, which naturally has favourable sampling properties (section 3.4), the accelerated acquisition was compared with standard breathold Cartesian 3D techniques for imaging arterial and venous phase imaging of contrast-enhanced liver MRI in eight participants. The same group has recently used similar techniques to provide rapid quantification of liver perfusion in patients undergoing clinical contrast enhancement (Chandarana et al 2015). Eightfold accelerated perfusion measurement during DCE-MRI has been demonstrated in the heart exploiting temporal redundancy (Otazo et al 2010).
5.6. Lung imaging with hyperpolarized gases
Breathable hyperpolarized gases such as helium-3 and xenon-129 have been used to provide high-resolution images of lung ventilation or function. The hyperpolarization provided is non-renewable as it is tipped into the transverse plane and used, and the patient must inhale the gas and hold their breath for high-quality imaging. Both of these considerations require that the imaging sequences used for 3He and 129Xe imaging must be rapid, and this is especially true if we wish to capture image series of gas inflow.
The Wild group at Sheffield has prospectively applied compressed sensing to accelerate 2D and 3D 3He imaging, by a factor of 2 in 2D imaging and by up to factor 5 in 3D imaging (Ajraoui et al 2010). Additionally, apparent diffusion coefficient, T2* and B1 maps were acquired in a single scan requiring a breath hold of less than 15 s (Ajraoui et al 2012). A further development was to use a proton image of the lungs acquired at the same time as a prior knowledge constraint in reconstructing the undersampled 3He images (Ajraoui et al 2013).
5.7. Spectroscopic imaging
Although the lower matrix sizes and SNR of 1H spectroscopic imaging would seem to weigh against compressed sensing, there are a growing number of studies using undersampling in this context which show promising results, such as (Sarma et al 2014). Compressed sensing may have a greater role to play in clinical spectroscopic imaging if the 13C hyperpolarized techniques with abundant SNR but limited polarization duration are translated to human studies: presently pre-clinical studies using compressed sensing have been demonstrated (Park et al 2013).
6. Alternative acceleration methods
As described in section 3, compressed sensing can be used to greatest advantage in situations where the k-space trajectory is 3D Cartesian, radial or spiral, where there is the opportunity to create undersampling patterns which produce non-aliased artefacts when subjected to inverse Fourier transform. Alternatively, several techniques have been developed which allow the simultaneous excitation of multiple 2D slices in the presence of a suitable phased array coil. These techniques have generally been termed 'simultaneous multi-slice imaging' or 'multiband', though there are a host of individual methods. This is particularly advantageous in neuro applications where there a preparation of magnetization is followed by an echo-planar readout, and where 32 channel head coils are being used with large numbers of small sensitive coils covering the anatomy. Such applications are common in diffusion-weighted measurements and in the acquisition of functional MRI of the brain (Chen et al 2015), and are playing a key role in the Human Connectome project. Preliminary work has been performed on diffusion tensor imaging of the heart (Lau et al 2015). The nature of the acquisitions being performed tends to indicate a 2D acquisition, which is often less favourable for high acceleration factors with compressed sensing. A detailed description of these techniques is beyond the scope of this article but the various techniques for simultaneous acquisition have been reviewed elsewhere (Feinberg and Setsompop 2013).
Another technique generating current research interest in that of magnetic resonance fingerprinting (Ma et al 2013). The fundamental idea is to create a very complex set of spin coherence states (using a balanced steady state free precession sequence) which give weightings of different image contrasts, and then to use a database to infer multiple reconstructed images from that state. For example, the reconstruction of proton density, T1 and T2 weighted brain images from a single set of coherence pathways has been demonstrated. The initial work of Ma suggests that fingerprinting may be robust to substantial spatial undersampling of the data. To date, there has been most interest in neuro applications, where spin evolution conditions and imperfections such as off-resonance can be controlled more closely (and where, for example, there is good separation of lipid and water structures). The field of MR fingerprinting is still at a preliminary stage and the roll-out to non-neuro applications and detailed validation studies remain to be done.
7. Future challenges
The use of undersampled data acquisition and compressed sensing reconstruction has the potential to reduce the total scan time and cost of MR acquisitions and to reduce the burden of imaging to patients. This can permit either greater patient throughput, or permit more imaging to be completed per unit time, which may be important for systemic metabolic and genetic diseases, where multiple organ systems are involved. Additionally, other scan types can be made feasible.
However, can we expect to see methods involving compressed sensing being used by radiologists in the foreseeable future, by clinical researchers only, or will this field be gently abandoned as a fashion? Despite extensive excellent work on MRI using radial and spiral geometries carried out over the last 20 years, there are almost no non-Cartesian pulse sequences in routine clinical use, and very few in practical clinical research, taking advantage of this work. Is compressed sensing doomed to the same fate? In the literature, a lack of translation to final users is presently discernable: while there are over 120 papers about compressed sensing in MRI published in Magnetic Resonance in Medicine, there are only 8 papers in Radiology.
Reasons for pessimism would be the inherent bias of research physicists to be naturally inclined to push on to the next innovation in accelerated imaging, rather than performing the necessary work to make compressed sensing practical for routine clinical use. A reason for optimism would be that the inherent sparsity of medical images is perhaps now one of the last sources of acceleration to be exploited. The field strength of MRI scanners being used has increased to 7 T or 3 T depending on application and we find ourselves increasingly constrained by physiological limitations of specific absorption rates and peripheral nerve stimulation. The author believes that compressed sensing will make a valuable contribution to the future of both clinical and research MRI. It will be necessary to remember that for all the increasingly complicated reconstructions that are proposed by physicists, it is essential for the radiographer to get image feedback within seconds of the scan terminating for accelerated imaging to be practically useful, and we must work within the reasonable computing power limits of the time. The second is that radiologists are presently happy to use acceleration techniques that can lead to artefacts (such as parallel imaging), provided that they understand the nature of the artefacts being produced. There are many unanswered questions in the field of MR image acceleration. How far will the techniques of multiband imaging come to dominate brain imaging, and can they find an important role in body imaging? Will Cartesian or radial compressed sensing come to be used as a clinical standard, given the increased computation requirements and general radiological reluctance to embrace radial imaging? Will the new field of MR fingerprinting make inroads into practical imaging in and even beyond the brain? It is certain, however, that these issues in compressed sensing will continue to call upon the skills and fire the imagination of MR physicists for the foreseeable future.
Acknowledgments
KGH gratefully acknowledges funding from the UK Medical Research Council (G1100160). The author would like to acknowledge the reviewers of this manuscript for their thoughtful criticism and useful suggestions.
Footnotes
- 1
For sequences with inversion preparation pulses, the definition of the repetition and inversion times can vary between vendors and acquisition time calculations need to take account of this (see McRobbie et al p 250).