Abstract
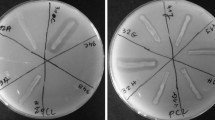
Four potential polyester-degrading bacterial strains were isolated from compost soils in Thailand. These bacteria exhibited strong degradation activity for polyester biodegradable plastics, such as polylactic acid (PLA), polycaprolactone (PCL), poly-(butylene succinate) (PBS) and polybutylene succinate-co-adipate (PBSA) as substrates. The strains, classified according to phenotypic characteristics and 16S rDNA sequence, belonging to the genera Actinomadura, Streptomyces and Laceyella, demonstrated the best polyester- degrading activities. All strains utilized polyesters as a carbon source, and yeast extract with ammonium sulphate was utilized as a nitrogen source for enzyme production. Optimization for polyester-degrading enzyme production by Actinomadura sp. S14, Actinomadura sp. TF1, Streptomyces sp. APL3 and Laceyella sp. TP4 revealed the highest polyester-degrading activity in culture broth when 1% (w/v) PCL (18 U/mL), 0.5% (w/v) PLA (22.3 U/mL), 1% (w/v) PBS (19.4 U/mL) and 0.5% (w/v) PBSA (6.3 U/mL) were used as carbon sources, respectively. All strains exhibited the highest depolymerase activities between pH 6.0–8.0 and temperature 40–60°C. Partial nucleotides of the polyester depolymerase gene from strain S14, TF1 and APL3 were studied. We determined the amino acids making up the depolymerase enzymes had a highly conserved pentapeptide catalytic triad (Gly-His-Ser-Met-Gly), which has been shown to be part of the esterase-lipase superfamily (serine hydrolase).
Similar content being viewed by others
References
Akutsu-Shigeno, Y., Teeraphatpornchai, T., Teamtisong, K., Nomura, N., Uchiyama, H., Nakahara, T., and Nakajima-Kambe, T., Cloning and sequencing of a poly(DL-lactic acid) depolymerase gene from Paenibacillus amylolyticus strain TB-13 and its functional expression in Escherichia coli, Appl. Environ. Microbiol., 2003, vol. 69, pp. 2498–2504.
Bradford, M.M., A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding, Anal. Biochem., 1976, vol. 72, pp. 248–254.
Calabia, B.P. and Tokiwa, Y., A novel PHB depolymerase from a thermophilic Streptomyces sp., Biotechnol. Lett., 2006, vol. 28, pp. 383–388.
Chua, T.K., Tseng, M., and Yang, M.K., Degradation of poly(ε-caprolactone) by thermophilic Streptomyces thermoviolaceus subsp. thermoviolaceus 76T-2, AMB Express, 2013, vol. 3, pp. 1–8.
Hanphakphoom, S., Maneewong, N., Sukkhum, S., Tokuyama, S., and Kitpreechavanich, V., Characterization of poly(L-lactide)-degrading enzyme produced by thermophilic filamentous bacteria Laceyella sacchari LP175, J. Gen. Appl. Microbiol., 2014, vol. 60, pp. 13–22.
Hu, X., Osaki, S., Hayashi, M., Kaku, M., Katuen, S., Kobayashi, H., and Kawai, F., Degradation of a terephthalate-containing polyester by thermophilic actinomycetes and Bacillus species derived from composts, J. Polym. Environ., 2008, vol. 16, pp. 103–108.
Hu, X., Thumarat, U., Zhang, X., Tang, M., and Kawai, F., Diversity of polyester-degrading bacteria in compost and molecular analysis of a thermoactive esterase from Thermobifida alba AHK119, Appl. Microbiol. Biotechnol., 2010, vol. 87, pp. 771–779.
Kieser, T., Bibb, M.J., Buttner, M.J., Chater, K.F., and Hopwood, D.A., Practical Streptomyces Genetics, Norwich: The John Innes Foundation, 2000.
Lane, D.J., 16S/23S rRNA sequencing, in Nucleic Acids Techniques in Bacterial Systematics, Stackebrandt, E. and Goodfellow, M., Eds., Wiley, 1991, pp. 115–147.
Li, F., Wang, S., Liu, W., and Chen, G., Purification and characterization of poly (L-lactic acid)-degrading enzymes from Amycolatopsis orientalis ssp. Orientalis, FEMS Microbiol. Lett., 2008, vol. 282, pp. 52–58.
Luengo, J.M., Garćia, B., Sandoval, A., Naharro, G., and Olivera, E.R., Bioplastics from microorganisms, Curr. Opin. Microbiol., 2003, vol. 6, pp. 251–260.
Matsuda, E., Abe, N., Tamakawa, H., Kaneko, J., and Kamio, Y., Gene cloning and molecular characterization of an extracellular poly(L-lactic acid) depolymerase from Amycolatopsis sp. strain K104-1, J. Bacteriol., 2005, vol. 187, pp. 7333–7340.
Mueller, R.J., Biological degradation of synthetic polyesters-enzymes as potential catalysts for polyester recycling, Proc. Biochem., 2006, vol. 41, pp. 2124–2128.
Nishida, H. and Tokiwa, Y., Distribution of poly(β-hydroxybutyrate) and poly(ε-caprolactone) aerobic degrading microorganisms in different environments, J. Environ. Polym. Degrad., 1993, vol. 1, pp. 227–233.
Oda, Y., Naoya, O., Teizi, U., and Kenzo, T., Polycaprolactone depolymerase produced by the bacterium Alcaligenes faeaclis, FEMS Microbiol. Lett., 1997, vol. 152, pp. 339–343.
Penkhrue, W., Khanongnuch, C., Masaki, K., Pathom-Aree, W., Punyodom, W., and Lumyong, S., Isolation and screening of biopolymer-degrading microorganisms from northern Thailand, World. J. Microbiol. Biotechnol., 2015, vol. 31, pp. 1431–1442.
Pranamuda, H., Yutaka, T., and Hideo, T., Polylactide degradation by Amycolatopsis sp., Appl. Environ. Microbiol., 1997, vol. 63, pp. 1637–1640.
Pridham, T.G., and Gottlieb, D., The utilization of carbon compounds by some Actinomycetales as an aid for species determination, J. Bacteriol., 1948, vol. 56, pp. 107–114.
Puhl, A.A., Selinger, L.B., McAllister1, T.A., and Inglis, G.D., Actinomadura keratinilytica sp. nov., a keratindegrading actinobacterium isolated from bovine manure compost, Int. J. Syst. Evol. Microbiol., 2009, vol. 59, pp. 828–834.
Saitou, N. and Nei, M., The neighbor-joining method: a new method for reconstructing phylogenetic trees, Mol. Biol. Evol., 1987, vol. 4, pp. 406–425.
Shimao, M., Biodegradation of plastics, Curr. Opin. Biotechnol., 2001, vol. 12, pp. 242–247.
Shinozaki, Y., Morita, T., Cao, X.H., Yoshida, S., Koitabashi, M., Watanabe, T., Suzuki, K., Sameshima-Yamashita, Y., Nakajima-Kambe, T., Fujii, T., and Kitamoto, H.K., Biodegradable plastic-degrading enzyme from Pseudozyma antarctica: cloning, sequencing, and characterization, Appl. Microbiol. Biotechnol., 2013. vol. 97, pp. 2951–2959.
Shirling, E.B. and Gottlieb, D., Methods for characterization of Streptomyces species, Int. J. Syst. Bacteriol., 1966, vol. 16, pp. 313–340.
Sriyapai, T., Somyoonsap, P., Matsui, K., Kawai, F., and Chansiri, K., Cloning of a thermostable xylanase from Actinomadura sp. S14 and its expression in Escherichia coli and Pichia pastoris, J. Biosci. Bioeng., 2011, vol. 111, pp. 528–536.
Staneck, J.L. and Roberts, G.D., Simplified approach to identification of aerobic actinomycetes by thin-layer chromatography, Appl. Microbiol., 1994, vol. 28, pp. 226–231.
Sukkhum, S., Tokuyama, S., Kongsaeree, P., Tamura, T., Ishida, Y., and Kitpreechavanich, V., A novel poly (L-lactide) degrading thermophilic actinomycetes, Actinomadura keratinilytica strain T16-1 and pla sequencing, Afr. J. Microbiol. Res., 2011, vol. 5, pp. 2575–2582.
Sukkhum, S., Tokuyama, S., and Kitpreechavanich, V., Poly(L-lactide)-degrading enzyme production by Actinomadura keratinilytica T16-1 in 3 L airlift bioreactor and its degradation ability for biological recycle, J. Microbiol. Biotechnol., 2012, vol. 22, pp. 92–99.
Suyama, T., Hosoya, H., and Tokiwa, Y., Bacterial isolates degrading aliphatic polycarbonates, FEMS Microbiol. Lett., 1998, vol. 161, pp. 255–261.
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S., MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods, Mol. Biol. Evol., 2011, vol. 28, pp. 2731–2739.
Techapun, C., Charoenrat, T., Watanabe, M., Sasaki, K., and Poosaran, N., Optimization of thermostable and alkaline-tolerant cellulose-free xylanase production from agricultural waste by thermotolerant Streptomyces sp. Ab106, using the central composite experimental design, Biochem. Eng. J., 2002, vol. 12, pp. 99–105.
Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F., and Higgins, D.G., The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools, Nucleic Acids Res., 1997, vol. 25, pp. 4876–4882.
Tokiwa, Y., and Calabia, B.P., Degradation of microbial polyesters, Biotechnol. Lett., 2004, vol. 26, pp. 1181–1189.
Tokiwa, Y., Iwamoto, A., Koyama, M., Kataoka, N., and Nishida, H., Biological recycling of plastics containing ester bonds, Makromol. Chem. Macromol. Symp., 1992, vol. 57, pp. 273–279.
Tseng, M., Yang, S.F., Hoang, K.C., Liao, H.C., Yuan, G.F., and Liao, C.C., Actinomadura miaoliensis sp. nov., a thermotolerant polyester-degrading actinomycete, Int. J. Syst. Evol. Microbiol., 2009, vol. 59, pp. 517–520.
Uchida, H., Nakajima-Kambe, T., Shigeno-Akutsu, Y., Nomura, N., Tokiwa, Y., and Nakahara, T., Properties of a bacterium which degrades solid poly(tetramethylene succinate)-co-adipate, a biodegradable plastic, FEMS Microbiol. Lett., 2000. vol. 189, pp. 25–29.
Author information
Authors and Affiliations
Corresponding author
Additional information
The article is published in the original.
Rights and permissions
About this article
Cite this article
Sriyapai, P., Chansiri, K. & Sriyapai, T. Isolation and Characterization of Polyester-Based Plastics-Degrading Bacteria from Compost Soils. Microbiology 87, 290–300 (2018). https://doi.org/10.1134/S0026261718020157
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0026261718020157