Soil Salinity Controls Relative Abundance of Specific Bacterial Groups Involved in the Decomposition of Maize Plant Residues
- 1Soil Ecology Lab, Cinvestav, Ciudad de México, Mexico, Mexico
- 2Cátedras CONACYT, Centro Tlaxcala de Biología de la Conducta, Universidad Autónoma de Tlaxacala, Tlaxcala, Mexico
Extreme salinity and alkalinity in soil is known to inhibit organic material decomposition and affect the bacterial community structure involved in its mineralization. Regular flooding of these soils will reduce salinity, which will alter the bacterial community involved in organic material mineralization. Soil of the former lake Texcoco with electrolytic conductivity (EC) 157.4 dS m−1 and pH 10.3 was flooded monthly, amended with maize plant residue or its neutral detergent fiber [NDF; mostly (hemi)cellulose and some lignin], while C mineralization and the bacterial community structure was monitored by means of 454 pyrosequencing of the 16S rRNA gene. The EC of the soil dropped from 157.8 to 1.7 dS m−1, but the pH (10.3) did not change significantly over time. On the one hand, the relative abundance of some bacterial groups, e.g., Bacillus and Gammaproteobacteria, always increased when maize plants or NDF were applied to soil independent of the changes in soil characteristics, i.e., they always participated in the degradation of the organic material applied, while the relative abundance of other groups, e.g., Acidobacteria, Alphaproteobacteria, Chloroflexi, Clostridia, Deltaproteobacteria, Gemmatimonadetes, Planctomycetes, and Verrucomicrobia, always decreased compared to the unamended soil. On the other hand, the increase or decrease of the relative abundance of other bacterial groups when organic material was applied to soil was influenced by the changes in soil characteristics. For instance, the relative abundance of the Actinomycetales, Halomonas, and Prauseria, did not increase when organic material was applied to soil with a high salt content, but did when the salt content was lowered while that of the Betaproteobacteria and Pirellulales increased when the salt content was high, but not when it was lowered. Application of the NDF generally had a similar effect on the bacterial community structure as when maize plants were applied. It was found that the capacity of some bacterial groups to degrade organic material was not affected by soil salt content, while that of others was stimulated or suppressed.
Introduction
Organic material plays an important role in soil, one of which is carbon sequestration, thereby mitigating global warming and is determinant in soil structure formation (Powlson et al., 2011; Stockmann et al., 2013; Coleman and Wall, 2014). When left on the soil surface, it prevents wind and water erosion and facilitates water infiltration. Most importantly, however, organic material serves as a C substrate for the soil microbial biomass and when mineralized provides nutrients for plants, e.g., N and P (Strickland et al., 2009; Dungait et al., 2012; Trivedi et al., 2013; Wieder et al., 2013). Application of organic material will affect the bacterial community structure in soil. The relative abundance of some bacteria will increase by organic material application (copiotrophs) while others will decrease (oligotrophs) in response to ecological succession or microbial substrate preferences (Goldfarb et al., 2011; Schimel and Schaeffer, 2012).
Salts concentration as a major stress to soil microorganisms has been subjected to several studies, where a decrease in CO2-evolution, enzymatic activity, or microbial biomass has often been observed (Sarig and Steinberger, 1994; Rietz and Haynes, 2003; Mamilov et al., 2004). Increasing salinity has a detrimental effect on biologically mediated processes in soil, such as C and N-mineralization (Pathak and Rao, 1998). In saline soils and under drought, microbes suffer from osmotic stress, which results in drying and lysis of cells (Oren, 2008), which reduces soil biomass.
Soil microorganisms have the ability to adapt to or tolerate osmotic stress caused by drought or salinity (Sardinha et al., 2003), they nevertheless have to cope with changing osmotic pressure and thus might change their physiology and morphology in response to this (Killham, 1994; Zahran, 1997). Killham (1994) describes two main adaptation strategies of microorganisms to osmotic stress (such as salinity, drought, or freezing), both result in accumulation of solutes in the cell to counteract the increased osmotic pressure. One strategy is to selectively exclude the incorporated solute (e.g., Na+, Cl−) and instead accumulate other ions necessary for metabolism (e.g., ). The other adaptation mechanism of the cell is to produce organic compounds, which will antagonize the concentration gradient between soil solution and cell cytoplasm. Some ions such as (Na+, Cl−) are exclusively transported trough the cell membrane, and other compounds are cell manufactured (betaine and malic acid), both are energy expensive strategies, however, those strategies were discover and have been studied in single microorganisms (Oren, 2002). These mechanisms are known to occur in individual microorganisms but have hardly ever been studied on a community level (Wichern et al., 2006).
Adverse effects of high salt concentrations in soils include dispersion and flocculation of soil particles that affect organic matter solubility and its availability, thus affecting the microbial community structure (Wong et al., 2008, 2009, 2010; Baumann and Marschner, 2013; Setia and Marschner, 2013). Little is known on how salinity affects soil microorganism communities and their metabolic capacities (Chowdhury et al., 2011; Rath and Rousk, 2015). It has been reported that salinity reduces the microbial biomass (Tripathi et al., 2006) and changes the microbial community composition (Wichern et al., 2006; Gennari et al., 2007; Chowdhury et al., 2011). Most of the studies consist of experiments where salts are added to soils (Rath and Rousk, 2015).
Soil from the former lake Texcoco is characterized by a high salinity and alkalinity. The Texcoco soil has been flooded since the mid 70's to lower the salt content so that it could be vegetated. The vegetation, initially the grass Distichlis spicata (L.) Greene and the salt cedar (Tamarix aphylla (L.) Karst.), prevents wind erosion and dust pollution in nearby Mexico city (Luna-Guido et al., 2000). Flooding the soil sharply decreased the salt content and altered the bacterial and archaeal population as determined by cloning and Sanger sequencing of the16S rRNA gene (Valenzuela-Encinas et al., 2008, 2009). Rath and Rousk (2015) stated that there is no current information on the effects of salinity on a natural saline soil microbial community using up-to-date molecular techniques, neither describing what would happen if this stress factor would be removed.
In a previous study, an extreme alkaline saline soil with pH 10.3 and EC 157.8 dS m−1 was flooded regularly for 10 months (de León-Lorenzana et al., 2017). By flooding the same soil samples under controlled conditions, the salt content decreased, while most other soil characteristics, e.g., pH, remained the same. Flooding the soil altered the bacterial community structure, but it remains to be investigated if the changes in salinity altered the bacterial metabolic capacity. Therefore, the soil was flooded, then amended with maize residue and its neutral detergent fraction [mostly (hemi) cellulose and some lignin] and incubated aerobically for 28 days. The C and N mineralization was determined during the aerobic incubation and the bacterial community structure monitored by means of 454 pyrosequencing of the 16S rRNA gene. The objective of this research was to study how lowering the soil salt content affected the bacteria involved in the degradation of organic material.
Materials and Methods
Soil Sampling Site
The soil from the former lake bed is located near the valley of Mexico City (Mexico) at an altitude of 2,240 masl with a mean annual temperature of 16°C and annual precipitation of 705 mm. Details of the soil characteristics and vegetation can be found in Luna-Guido et al. (2000). Soil was sampled at random with a stony soil auger diameter seven cm (Eijkelkamp, Giesbeek, Nl) by augering 30-times the 0–15 cm layer of three different plots with a size of ca. 400 m2. As such, approximately 30 kg soil was collected from each plot. The soil from each plot was pooled (so that three soil samples were obtained) and taken to the laboratory. This field-based replication was maintained in the laboratory experiment.
Flooding of the Soils and Incubation of the Columns
The soil from each plot (n = 3) was passed separately through a 5 mm sieve, adjusted to 40% water holding capacity (WHC) and was pre-incubated separately in drums for 7 days. Each drum with a 70 l volume capacity contained 10 kg soil and was closed airtight. The drums contained a beaker with 500 ml distilled H2O to avoid desiccation and a beaker with 1,000 ml 1 M NaOH solution to trap CO2 evolved. The drums were opened every day to avoid anaerobic conditions in the soil.
After 7 days, 8 sub-samples of 1.5 kg soil from each plot (n = 3) were packed separately into PVC columns with diameter 10.5 cm and length 30 cm to a bulk density of 0.95 g cm−3, the mean bulk density found in this soil (Beltrán-Hernández et al., 2007). The columns were fitted at the bottom with thin plastic plates with holes drilled equally distributed over their surface. On top of it, a polyethylene filter disc (nominal pore size 0.5 μm) was placed and a layer of acid washed sand 0.16 kg to prevent loss of soil particles during leaching (Bellini et al., 1996).
The experimental design was complex and therefore given in Figure S1. One column from each plot (n = 3) was selected at random and the soil was removed. Five hundred g were used to characterize the soil (Table 1) while the rest was used for an aerobic incubation experiment described below.
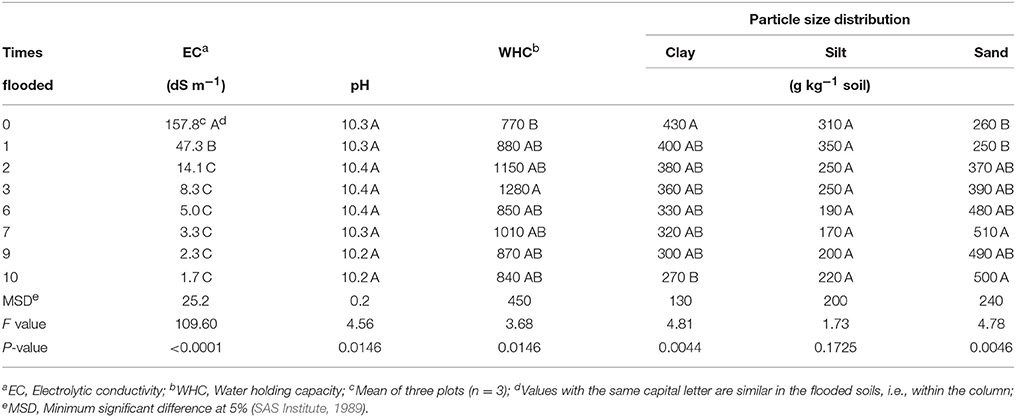
Table 1. Characteristics of soil of the former Lake Texcoco that was never flooded and soil flooded monthly, drained freely and incubated at approximately 50% of water holding capacity monthly for 10 months.
The remaining columns were flooded with 3 l distilled water and drained freely until approximately 50% WHC. The top of the columns were fitted with parafilm to avoid drying of the soil but to allow aeration. The soil was then conditioned aerobically at constant water content (approximately 50% WHC) for 1 month. Distilled water was applied to the soil columns when required to maintain the soil at 50% WHC. After 1 month, the soil was removed from three columns, three different treatments were applied and the soil was incubated aerobically as described below.
This process of flooding the soil, draining the soil freely until ca. 50% WHC, covering the column with parafilm and conditioning the soil for a month was repeated until the soil was flooded ten times, i.e., kept for a total of 10 months. Previous experiments with the soil from Texcoco showed that 8 months of monthly flooding was sufficient to markedly decrease the EC (Dendooven et al., 2015). After 2, 3, 6, 7, 9, and 10 floodings, one column was selected at random from each plot (n = 3). The soil was removed from the column, and three different treatments were applied and the soil was incubated aerobically.
Cultivation of the Maize Plants
The same maize plants were used in this experiment as in the one used in the study reported by Ramírez-Villanueva et al. (2015). Briefly, maize seeds were surface-sterilized and germinated on 0.8% agar-water plates. The maize seedlings with roots of 2 cm were placed on sterilized and C-free vermiculite in an acrylic growth chamber (105 l, 35 × 50 and 60 cm high) and moistened regularly with a nutritive Steiner solution (1961). After 25 days, the maize plants were harvested, air-dried, and characterized.
The maize plants were fractionated to obtain the neutral detergent fiber (NDF) fraction (Van Soest, 1963; Van Soest and Wine, 1967). Details of the fractionation of the maize plants can be found in Ruíz-Valdiviezo et al. (2010). A brief hot extraction with neutral detergent solution was used to remove the “soluble” part of the maize residue, leaving the NDF fraction containing most of the cell wall constituents, i.e., (hemi) cellulose plus some lignin (Table S1).
Aerobic Incubation
The unflooded and, once, twice, thrice, six times, seven times, nine times, and ten times flooded soils were used in the aerobic incubation experiment. Three different treatments were applied to the unflooded and flooded soil.
Twelve sub-samples of 30 g soil from, each plot (n = 3) and soil never flooded, the soil flooded once, twice, three times, six times, seven times, nine times, and ten times (n = 8) were added separately to 120 ml glass flasks. Four sub-samples soil were amended with maize residue at 2 g C kg−1 soil and four with the NDF fraction at 2 g C kg−1 soil. The remaining four soil samples were left unamended and served as control. One flask was chosen at random from the soil amended with maize residue, one amended with the NDF fraction and one left unamended, i.e., time 0. The soil was removed from the flasks and kept at−70 °C pending DNA extraction.
The remaining glass flasks were placed separately in 945 ml glass jars containing a vessel with 10 ml distilled H2O and one with 20 ml 1 M NaOH. The jars were sealed and stored in the dark at 22 ± 1°C for 28 days. An additional 15 jars containing a vessel with 10 ml distilled H2O and one with 20 ml 1 M NaOH were sealed and served as the control to account for the CO2 trapped from the atmosphere. After 7, 14, and 28 days, one jar was selected at random with soil amended with maize residue or the NDF fraction or left unamended, opened and the vessel containing NaOH removed. An aliquot of 5 ml of the 1 M NaOH was taken to determine CO2 trapped (Jenkinson and Powlson, 1976). The soil sample was removed from the glass flask and kept at −70°C pending DNA extraction. All remaining flasks were opened, aired for 10 min to avoid anaerobic conditions, re-sealed and further incubated.
Soil Characterization
The pH was determined in 1:2.5 soil–H2O suspension using a calibrated Ultra Basic UB-10 pH/mV meter (Denver Instrument, NY, USA) with a glass electrode (#3007281 pH/ATC). The EC was measured with a portable microprocessor HI 933300 (HANNA Instruments, Woonsocket, Rhode Island, USA). The WHC was measured by using 50 g dry soil sample which was placed in a funnel, water-saturated, covered with an aluminum foil to avoid water evaporation and left to stand overnight to drain freely. The WHC was defined as the amount of water retained in the water-saturated soil left to stand overnight. The soil particle size distribution was determined by the hydrometer method as described by Gee and Bauder (1986).
DNA Extraction and PCR Amplification of Bacterial 16S rRNA Genes
DNA was extracted from each soil sample as previously described by Ramírez-Villanueva et al. (2015). First, humic and fulvic acids were removed with pyrophosphate and three different cell lysis techniques were applied (Hoffman and Winston, 1987; Sambrook and Russell, 2001; Valenzuela-Encinas et al., 2008) to obtain an optimum representation of all the microorganisms in the soil samples (Carrigg et al., 2007). Each technique was used to extract DNA from 0.5 g soil twice (a total 1 g soil) and pooled. As such, DNA was extracted from 3 g soil per plot (n = 3), so overall 9 g soil DNA was extracted per treatment and incubation time. The precipitation of the proteins and purification of the DNA are described in Valenzuela-Encinas et al. (2008).
The V1-V3 region of bacterial 16S rRNA gene was amplified with 10-pb tagged primers 8-F (5′-AGA GTT TGA TCI TGG CTC A-3′) and 556-R 5′-TGC CAG IAG CIG CGG TAA-3′) containing the 454 FLX adapters (Navarro-Noya et al., 2013). The PCR reactions were done as previously described by Navarro-Noya et al. (2013). The product of five reactions of each metagenomic DNA sample was pooled to minimize PCR bias (Acinas et al., 2004) and constituted a single library. All the pyrosequencing libraries were purified using the DNA Clean & Concentrator purification kit as recommended by the manufacturer (Zymo Research, Irvine, CA, USA), and quantified using the PicoGreen® dsDNA assay (Invitrogen, Carlsbad, USA) and the NanoDropTM 3300 Fluorospectrometer (Thermo Scientific NanoDrop). Sequencing was done by Macrogen Inc. (DNA Sequencing Service, Seoul, Korea) by using a Roche 454 GS-FLX Titanium System pyrosequencer (Roche, Mannheim, Germany). All the pyrosequencing–derived 16S rRNA gene sequence datasets were submitted to the NCBI Sequence Read Archive (SRA) under accession number BIOPROJECT PRJNA308500, SRR4298851-SRR4298883, SRR3992369-SRR3992416, SRR3992178-SRR3992250, SRR3991784-SRR3991784.
Analysis of Pyrosequencing Data
The QIIME version 1.9.0 software was used to analyse the pyrosequencing data (Caporaso et al., 2010a). Different pipelines to analyse DNA sequences have been developed, e.g., mothur, QIIME, BMP, Kraken, Clark, and one Codex (Siegwald et al., 2017). We decided to use QIIME (version 1.9) as it is used widely, easy to determine alpha and beta diversity indices and contains tools for comparing phylogenetic information within the pipeline (Nilakanta et al., 2014). In the first step, poor quality reads were eliminated from the data sets, i.e., quality score <25, containing homopolymers > 6, length <400 nt, and containing errors in primers and tags. Denoising of the reads was done with the script denoise wrapper.py using the barcode-sorted libraries and the standard flowgram format (SFF) (Reeder and Knight, 2010). Operational taxonomic units (OTU) were determined at a 97% similarity level (OTU-97%) with UCLUST algorithm (Edgar, 2010) and one representative sequence of each OTU-97% was selected, i.e., rep-set. Chimeras were detected and removed from the rep-set using the Chimera Slayer (Haas et al., 2011). Sequence alignments of the rep-set were done against the Greengenes core set using PyNAST and filtered at a threshold of 75% (OTU-75%) (Caporaso et al., 2010b).
Phylogenetic and Statistical Analysis
The taxonomic distribution estimates at different levels were done using the taxonomy assignation at a confidence threshold of 80% by the naïve Bayesian rRNA classifier from the Ribosomal Data Project (http://rdp.cme.msu.edu/classifier/classifier.jsp) (Wang et al., 2007).
Diversity and species richness estimators were calculated from a 2,700 sequence rarified table to avoid bias due to the differences in the size of the data sets. A maximum-likelihood phylogenetic tree was constructed with the aligned sequences using FastTree 2.1.3 (Price et al., 2009) within QIIME. A UniFrac distance matrix was generated using the phylogenetic information and occurrence data to compare bacterial communities in the different treatments. Analysis of similarities (ANOSIM) and Permutational Multivariate Analysis of Variance (PERMANOVA) was done using unweighted UniFrac pairwise distances, to test significant differences between bacterial communities from the different treatments (n = 999). Soil characteristics were subjected to a one-way analysis of variance using PROC GLM (SAS Institute, 1989) to test for significant differences between the flooded soils with the Tukey's Studentized Range test.
Relative abundances of the different phyla, orders and genera were separately explored with a PCA using PROC FACTOR (SAS Institute, 1989). A canonical correlation analysis (CCA) was used to study the degree of relationship between the abundance of the different phyla, order and genera and the soil characteristics. The CCA was done using the PROC CANCORR of the SAS statistical package (SAS Institute, 1989). BioEnv analyses were used to assess how well the community structure was explained by environmental variables using non-factorial metadata. For BioEnv analyses the Vegan package in R (R Development Core Team, 2008) was used to create distances matrices of soil characteristics (Euclidean distances), i.e., pH, EC, WHC, and clay and sand content, and community composition (Unifrac distances), which were then compared through Spearman's rank coefficients (https://www.rdocumentation.org/packages/vegan/versions/2.4-2/topics/bioenv) (Clarke and Ainsworth, 1993).
The effect of flooding the soil on the relative abundance of the different bacteria groups was expressed as:
Ratio = [(relative abundance of the bacterial group in the flooded soil—relative abundance of the bacterial group in the unflooded soil)/ relative abundance of the bacterial group in the unflooded soil] (Equation 1).
Heatmaps with the ratios were made in R programming language using the libraries pheatmap and gplots (R Development Core Team, 2008). The ratios were categorized before the heatmap was constructed with value −5: ratio between the flooded and unflooded soil ≤ −0.99, value −4: ratio > −0.99 ≤ −0.96, value −3: ratio > −0.96 and ≤ −0.80, value −2: ratio > −0.80 ≤ −0.50, value −1: ratio > −0.50 > 0, value 0: ratio = 0, value 1: ratio > 0 and < 1, value 2: ratio ≥ 1 and < 4, value 3: ratio ≥ 4 and < 24, value 4: ratio ≥ 24 and < 99, value 5: ratio ≥ 99.
The effect of the application of maize plantlets or the NDF fraction on the relative abundance of the different bacterial groups was expressed as:
Ratio = [(relative abundance of the bacterial group in the maize amended soil—relative abundance of the bacterial group in the unamended soil) / relative abundance of the bacterial group in the unamended soil] (Equation 2).
Heatmaps with the ratios were made in R programming language using the libraries pheatmap and gplots (R Development Core Team, 2008). The ratios were categorized before the heat map was constructed with value −5: ratio between the flooded and unflooded soil ≤ −0.99, value −4: ratio > −0.99 ≤ −0.96, value −3: ratio > −0.96 and ≤ −0.80, value −2: ratio > −0.80 ≤ −0.50, value −1: ratio > −0.50 > 0, value 0: ratio = 0, value 1: ratio > 0 and < 1, value 2: ratio ≥ 1 and < 4, value 3: ratio ≥ 4 and < 24, value 4: ratio ≥ 24 and < 99, value 5: ratio ≥ 99.
Results
Soil Characteristics
The EC of the Texcoco soil dropped rapidly. After 3 floodings it was only 8.3 dS m−1 and after 10 floodings 1.7 dS m−1 (Table 1). The flooding reduced the clay particles from 430 g kg−1 at the onset of the experiment to 270 g kg−1 after 10 floodings. The WHC showed a lot of variation but was not affected significantly by the number of floodings. Changes in pH with flooding were minimal with a mean pH of 10.3.
Alpha Diversity
Overall 631,155 non-chimeric, good quality sequences were retrieved from the soil representing 8917 OTUs (OTU-97%). The number of sequences retrieved was sufficient as the rarefaction curve was asymptotic in the unamended or soil amended with maize plants or NDF (Figure S2). Flooding reduced the species richness significantly, i.e., Chao1 index, but did not affect the diversity, i.e., Simpson, Shannon, and the phylogenetic indices (Table S2). Overall, phylotypes belonging to 29 phyla, 88 classes, 165 orders, 309 families, and 510 genera were retrieved from the soil.
Bacterial Population Structure in the Unamended Flooded Soils
The relative abundance of the different bacterial groups showed large fluctuations in the unflooded soil incubated for 28 days (Figure S3). The relative abundance of the Proteobacteria, for instance, dropped from 83.4% at day 0 to 7.2% at day 7, while that of the Firmicutes (mostly Bacillales) increased from 2.7 to 31.6%. These fluctuations in relative abundance of different bacterial groups during the 28-day aerobic incubation decreased with increased flooding. Approximately 25% of the retrieved sequences remained unassigned independent of the number of floodings.
Flooding the soil affected the relative abundance (mean of the 28-day incubation) of different bacterial groups. For instance flooding the soil once increased the relative abundance of the Actinobacteria, Gemmatimonadetes, and [Thermi] compared to the unflooded soil, but further increases were slight while that of the Planctomycetes, TM7 and Verrucomicrobia tended to increase further with increased flooding (Figure 1). The relative abundance of Acidobacteria, Cyanobacteria, Chlorobi, Firmicutes, SBR1093, and Spirochaetes decreased in the flooded soil compared to the unflooded soil.
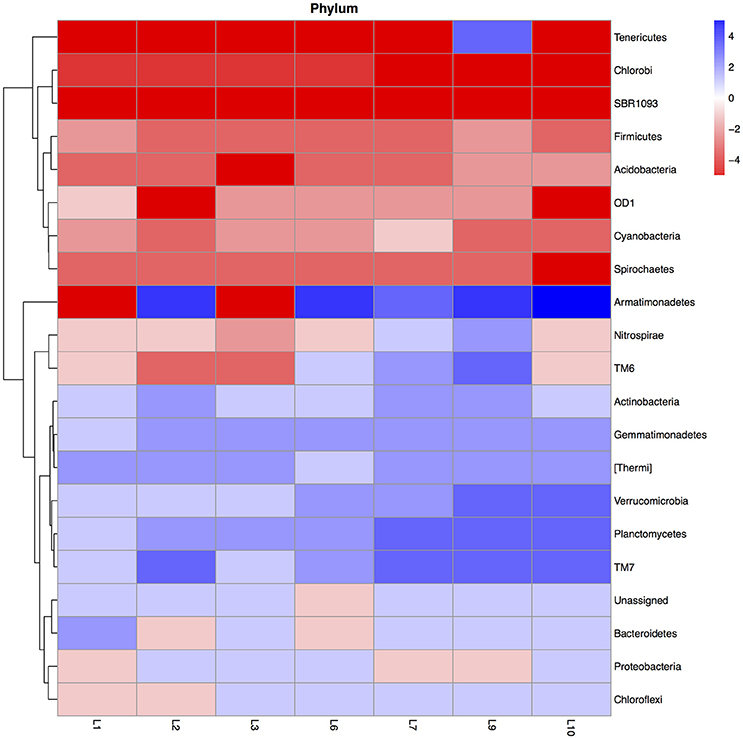
Figure 1. Heatmap of the ratio between the relative abundance of the different bacterial phyla in the unflooded soil vs. that of the soil flooded once (1), twice (2), trice (3), six (6), seven (7), nine (9), or ten times (10). The ratio was calculated as: (relative abundance of the bacterial group in the flooded soil—relative abundance of the bacterial group in the unflooded soil)/relative abundance of the bacterial group in the unflooded soil. The ratios were categorized with value −5: ratio between the flooded and unflooded soil ≤ −0.99, value −4: ratio > −0.99 ≤ −0.96, value −3: ratio > −0.96 and ≤ −0.80, value −2: ratio > −0.80 ≤ −0.50, value −1: ratio > −0.50 > 0, value 0: ratio = 0, value 1: ratio > 0 and < 1, value 2: ratio ≥ 1 and < 4, value 3: ratio ≥ 4 and < 24, value 4: ratio ≥ 24 and < 99, value 5: ratio ≥ 99.
The PCA visualized these changes clearly (Figure 2). First, the relative abundance of different bacterial groups showed large variations in the unflooded soil during the 28-day incubation. The bacterial community structure in the unflooded soil at day 7 and 14 was clearly different from that at day 0 and 28. Second, the biggest change in bacterial community structure was after just one flooding while changes with subsequent flooding were smaller. The unflooded soil was characterized mostly by a positive PC1 (larger relative abundance of the Firmicutes and Acidobacteria) and a negative PC2 (larger relative abundance of the Proteobacteria), while flooding favored Actinobacteria, Gemmatimonadetes, Planctomycetes, [Thermi], TM7, and Verrucomicrobia.
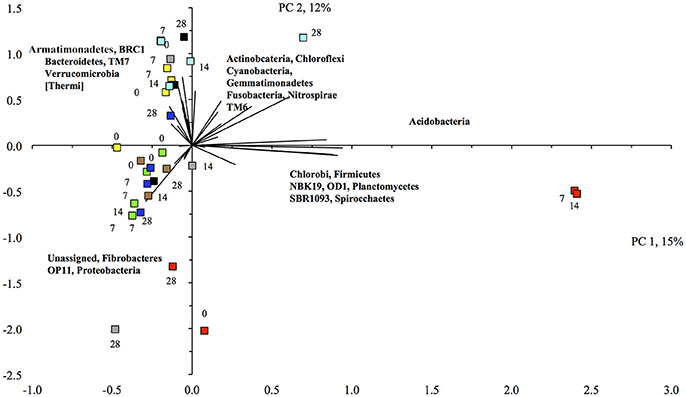
Figure 2. Principal component analysis with the different bacterial phyla retrieved from the unamended unflooded soil ( ), flooded once (
), flooded once ( ), twice (
), twice ( ), trice (
), trice ( ), six (
), six ( ), seven (
), seven ( ), nine (
), nine ( ), or ten times (
), or ten times ( ).
).
The CCA analysis indicated that most soils flooded > 3 times were grouped in the lower left quadrant, i.e., higher sand content and WHC and higher relative abundance of Actinobacteria, Gemmatimonadetes, and Verrucomicrobia, and the majority of soils flooded <3 times mostly in the upper and lower left quadrants, i.e., higher relative abundance of Acidobacteria, Bacteroidetes, and Proteobacteria, and a higher EC (Figure 3). However, the variation between some of the replicated soil samples was great, e.g., the soil flooded three, six and ten times. The BioEnv analysis showed that the bacterial community in the unamended soil correlated positively with clay content with a Spearman's rank correlation coefficient of 0.438.
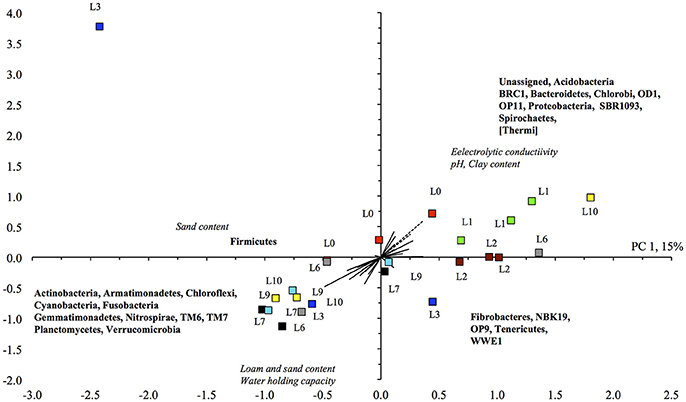
Figure 3. Canonical component analysis with the different bacterial phyla retrieved from the unamended unflooded soil ( ), flooded once (
), flooded once ( ), twice (
), twice ( ), trice (
), trice ( ), six (
), six ( ), seven (
), seven ( ), nine (
), nine ( ), or ten times (
), or ten times ( ) at time 0 and the different soil characteristics.
) at time 0 and the different soil characteristics.
C Mineralization
Flooding the soil once sharply decreased the CO2 emitted, but further flooding had no clear effect (Figure 4). Application of maize plants and NDF increased the CO2 emitted significantly, independent of the number of floodings (P < 0.05). The difference between the CO2 emitted from the NDF amended soil and the unamended soil increased from 84 mg C kg−1 soil (or 4% of the added organic material considering no priming effect) in the unflooded soil to 499 mg C kg−1 in the soil flooded twice, and decreased to 243 mg C kg−1 soil in soil flooded six times. It remained constant thereafter. A similar pattern was found in the maize plants amended soil, i.e., the difference between the maize amended soil and the unamended soil was 353 mg C kg−1 (or 18% of the added organic material considering no priming effect) in the unflooded soil, 836 mg C kg−1 in the soil flooded twice, 549 mg C kg−1 in soil flooded six times and remained constant thereafter.
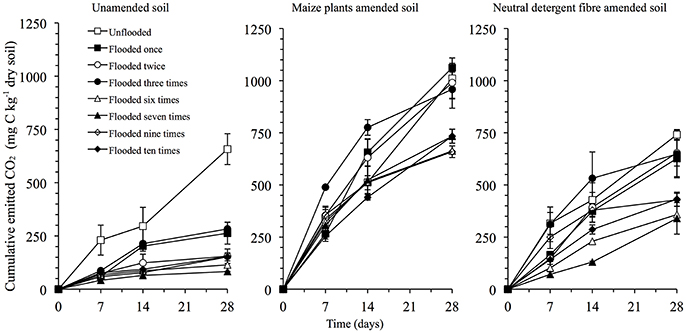
Figure 4. Emission of CO2 (mg C kg−1 dry soil) from the unamended soil and soil amended with maize plants or its neutral detergent fraction.
Bacterial Population in the Maize Residue and NDF Amended Flooded Soils
Application of NDF or maize changed the bacterial community structure and the changes in the relative abundance of the bacterial groups when the organic material was applied were affected by the number of floodings. Fluctuations over time in the 28-day incubation were larger in the unflooded soil amended with maize or NDF than in the soil flooded 10 times amended with organic material (Figures S4, S5). Application of maize plants or NDF to the unflooded soil increased the relative abundance of the Firmicutes (Bacillales and Oceaonospirillales) and the relative abundance of the Actinobacteria (mostly Actinomycetales) when the soil was flooded ten times (Figures S4, S5).
The relative abundance of only a limited number of bacterial groups increased when maize or NDF was applied to soil compared to the unamended soil (Figure 5, Figure S6). For instance, considering only the bacterial phyla, then the relative abundance of the Firmicutes increased mostly when maize plantlets and sometimes when NDF was applied to the flooded soil. Although the relative abundance of the Actinobacteria decreased when NDF and maize were applied to the unflooded soil (P < 0.0001), this trend was reversed after the soil was flooded.
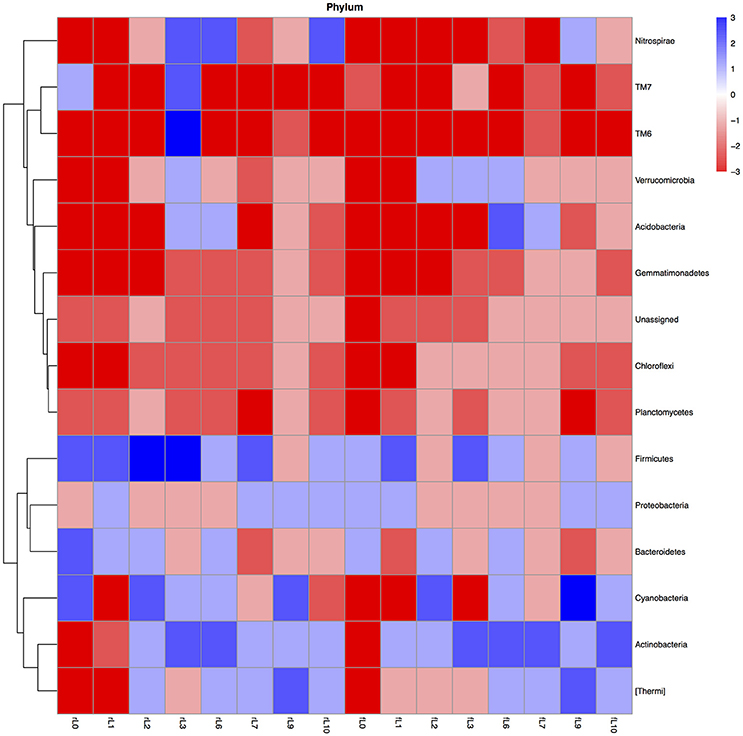
Figure 5. Heatmap of the ratio between the relative abundance of the different bacterial phyla in the unamended soil vs. the soil amended with maize plantlets or its neutral detergent fiber (NDF) for soil never flooded (rL0) flooded once (rL1), twice (rL2), trice (rL3), six (rL6), seven (rL7), nine (rL9), or ten times (rL10). The ratio was calculated as: [(relative abundance of the bacterial group in the maize amended soil—relative abundance of the bacterial group in the unamended soil)/relative abundance of the bacterial group in the unamended soil]. Heatmaps with the ratios were made in R programming language using the libraries pheatmap and gplots. The ratios were categorized with value −5: ratio ≤ −0.99, value −4: ratio > −0.99 ≤ −0.96, value −3: ratio > −0.96 and ≤ −0.80, value−2: ratio > −0.80 ≤ −0.50, value −1: ratio > −0.50 > 0, value 0: ratio = 0, value 1: ratio > 0 and < 1, value 2: ratio ≥ 1 and < 4, value 3: ratio ≥ 4 and < 24, value 4: ratio ≥ 24 and < 99, value 5: ratio ≥ 99.
Application of maize or NDF to the flooded soils decreased the relative abundance of a larger number of bacterial groups (Figure 5, Figure S6). The relative abundance of Chloroflexi, Gemmatimonadetes, Planctomycetes, and the unassigned sequences nearly always decreased when maize or NDF were applied.
The Anosim and PERMANOVA analyses of the bacterial populations in the three different treatments (unamended soil, and maize and NDF amended soil) showed that bacterial population in the unamended soil was always very significantly different to the maize and NDF amended soil. The bacterial community in the maize amended soil was significantly different to the NDF amended soil with EC 157.8, 47.3, 5.0, and 3.3 dS m−1 (Table S3).
The PCA showed a clear effect of flooding on the bacterial community structure of the maize-amended soil (Figure 6). The unflooded soil was located in the upper left quadrant, i.e., with the highest relative abundance of the Bacteroidetes and Firmicutes compared to other soils, and a low relative abundance of Actinobacteria, Gemmatimonadetes, Planctomycetes, and Verrucomicrobia. The soil flooded once was also located in the upper left quadrant, but the value for PC2 was lower, i.e., a lower relative abundance of Bacteroidetes and Firmicutes, than in the unflooded soil. Flooding the soil more increased the relative abundance of the Actinobacteria, Gemmatimonadetes, Planctomycetes, and Verrucomicrobia, i.e., the value of PC1 increased. Consequently, the soils flooded nine and ten times were found in the lower right quadrant, i.e., they had positive PC1 or the highest relative abundance for the Actinobacteria, Gemmatimonadetes, Planctomycetes, and Verrucomicrobia.
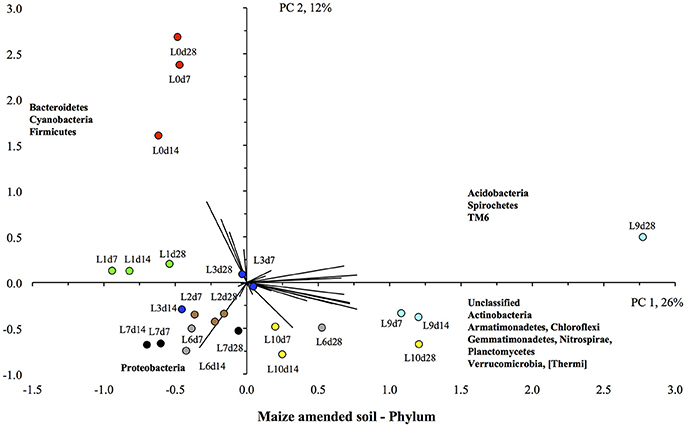
Figure 6. Principal component analysis with the different bacterial phyla retrieved from the unflooded soil amended with maize plants ( ), or flooded once (
), or flooded once ( ), twice (
), twice ( ), trice (
), trice ( ), six (
), six ( ), seven (
), seven ( ), nine (
), nine ( ), or ten times (
), or ten times ( ).
).
The PCA also showed a clear effect of flooding on the bacterial community structure of the NDF-amended soil (Figure 7). The unflooded soil was located in the lower left quadrant, i.e., with the highest relative abundance of the Bacteroidetes, Firmicutes, and Proteobacteria compared to other soils, and a low relative abundance of Actinobacteria, Gemmatimonadetes, Planctomycetes, and Verrucomicrobia. The soils flooded once and twice were still located in the lower left quadrant, but the value for PC1 was lower, i.e., a lower relative abundance of Bacteroidetes and Firmicutes, than in the unflooded soil. Flooding the soil more increased the relative abundance of the Actinobacteria, Gemmatimonadetes, Planctomycetes, and Verrucomicrobia, i.e., the value of PC1 increased. Thus, the soils flooded nine and ten times were found in the upper and lower right quadrant, i.e., they had positive PC1 or the highest relative abundance for the Actinobacteria, Gemmatimonadetes, Planctomycetes, and Verrucomicrobia.
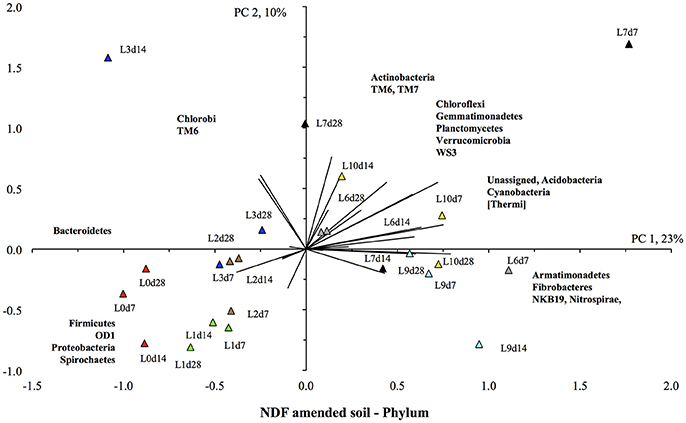
Figure 7. Principal component analysis with the different bacterial phyla retrieved from the unflooded soil amended with the neutral detergent fiber fraction ( ), or flooded once (
), or flooded once ( ), twice (
), twice ( ), trice (
), trice ( ), six (
), six ( ), seven (
), seven ( ), nine (
), nine ( ), or ten times (
), or ten times ( ).
).
The spearman rank coefficient (r) between the soil bacterial communities and physicochemical soil properties was high. The BioEnv analysis showed that the community in the NDF treatment correlated with EC, silt and sand content, and in the Maize treatment with EC, with a Spearman's rank correlation coefficient of 0.863 and 0.922, respectively.
Degraders of the Organic Material Applied
Some distinctive patterns explain how bacterial groups respond to the application of maize plants to soil with a high pH when the salt content decreased due to flooding (Table 2, Figures 5, 8–11). First, some bacterial groups were not favored (their relative abundance decreased) by the application of organic material, and changes in the ratio of the relative abundance in the unamended soil vs. the maize amended soil were small or showed no clear pattern with decreasing salt content. They can be considered “Undefined.” Different bacterial groups, such as the Bacteroidetes, Marinimicrobium, and Nitriluptor, showed no clear pattern in the ratio of unamended soil vs. the maize amended soil. The ratio in the relative abundance of the maize amended soil vs. the unamended soil of phylotypes belonging to the Proteobacteria also had no clear pattern with decreasing EC although at lower taxonomic levels of this phylum, bacterial groups were clearly favored by the application of the maize plants and their response was related often to the salt content of the soil. Second, some bacterial groups were favored by the application of the maize plants and they participated in its decomposition independent of the soil salt content. They can be considered “Copiotrophs.” The relative abundance of phylotypes belonging to bacterial groups, such as Bacillus, always increased in the maize plants amended soil compared to the unamended soil. Third, other bacterial groups were favored by the application of the maize plants but only after the soil was flooded and the EC decreased. They participated in the degradation of the maize plants but only when the salt content had decreased substantially, so they can be considered “Saline adverse copiotrophs.” The relative abundance of phylotypes belonging to bacterial groups, such as Nocardiopsis, Prauseria, and Streptomyces, increased in the maize plants amended soil compared to the unamended soil when the soil was flooded at least once. Fourth, application of maize plants favored little bacterial groups but decreasing the EC inhibited them. They participated in the degradation of the maize plants when the salt content was high but not when the salt content was low and can be considered “Saline copiotrophs.” They were only a small group and the relative abundance of phylotypes belonging to bacterial groups, such as Idiomarinaceae, Sphingobacteriales, was affected in this way. Fifth, the relative abundance of some bacterial groups decreased when maize plant was applied independently of changes in salt content. They can be considered as “Oligotrophs.” The relative abundance of phylotypes belonging to these bacterial groups, such as Deltaproteobacteria, Gemm-5, and Planctomycetes, decreased in the maize plants amended soil compared to the unamended soil independent of the soil salt content. Sixth, the relative abundance of these bacterial groups decreased strongly when maize plants were applied to the soil, but the inhibitory effect decreased when the soil salt content decreased with flooding. They can be considered “Saline oligotrophs.” The relative abundance of phylotypes belonging to bacterial groups, such as Acididomicrobiales, Alphaproteobacteria, Chloroflexi, Euzebya, Gemmatimonadetes, and Rubricoccus Verrucomicrobia, decreased in the maize plants amended soil compared to the unamended soil independent of the soil salt content.
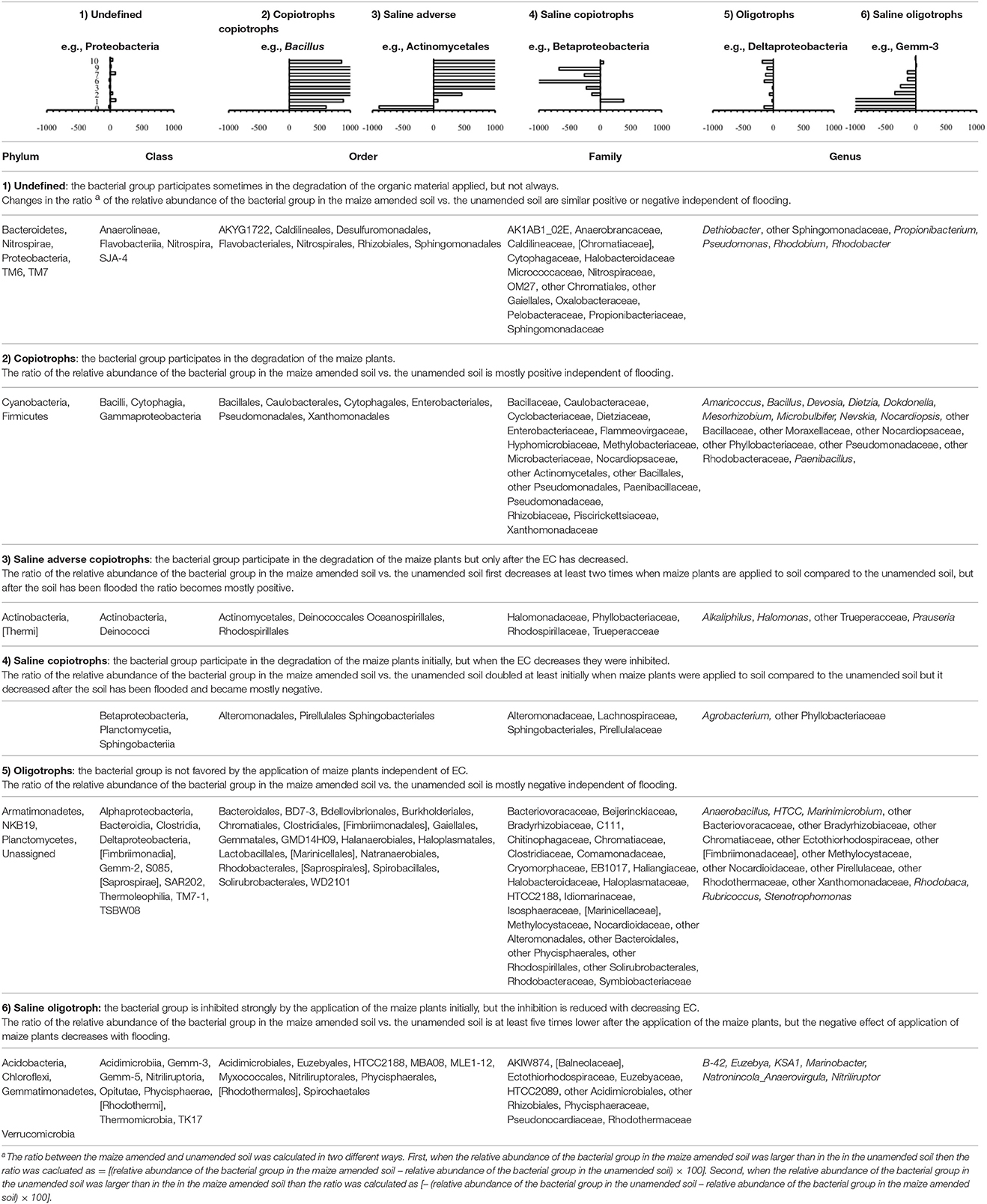
Table 2. Effect of decreasing electrolytic conductivity (EC) of the Texcoco soil due to flooding and its effect on the ratio of the relative abundance of bacterial group in the unamended soil vs. the maize-amended soil.
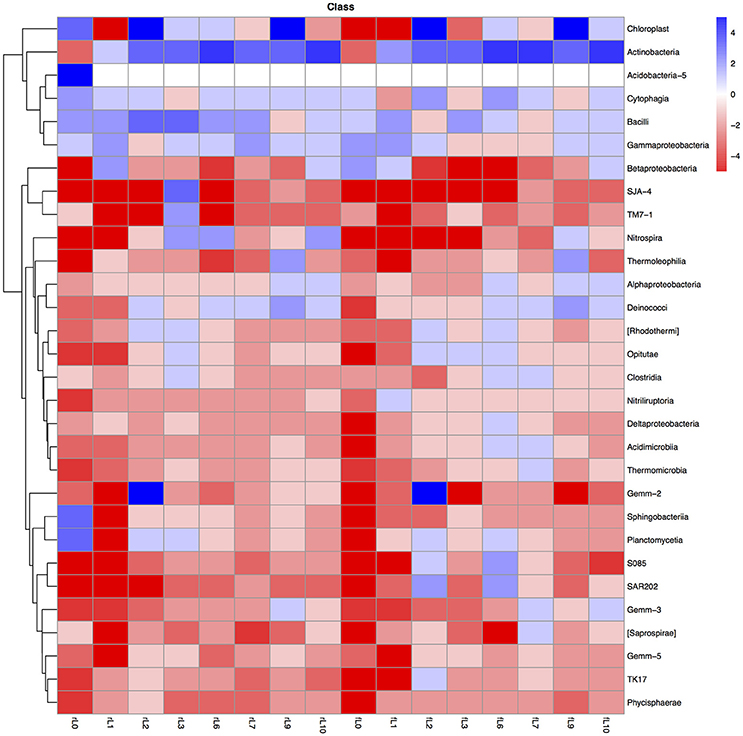
Figure 8. Heatmap of the ratio between the relative abundance of the different bacterial classes in the unamended soil vs. the soil amended with maize plantlets or its neutral detergent fiber (NDF). Legends to the figure can be found in Figure 5.
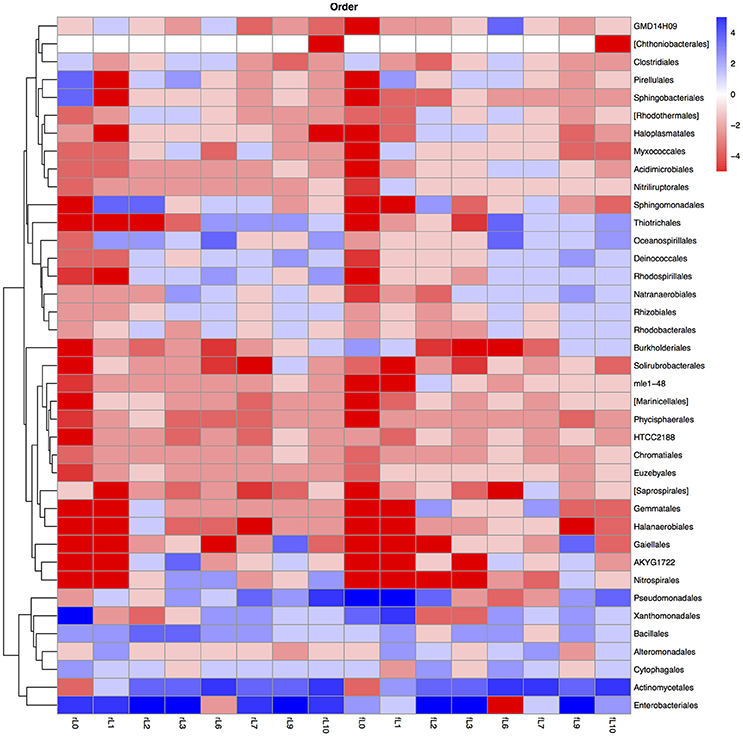
Figure 9. Heatmap of the ratio between the relative abundance of the different bacterial orders in the unamended soil vs. the soil amended with maize plantlets or its neutral detergent fiber (NDF). Legends to the figure can be found in Figure 5.
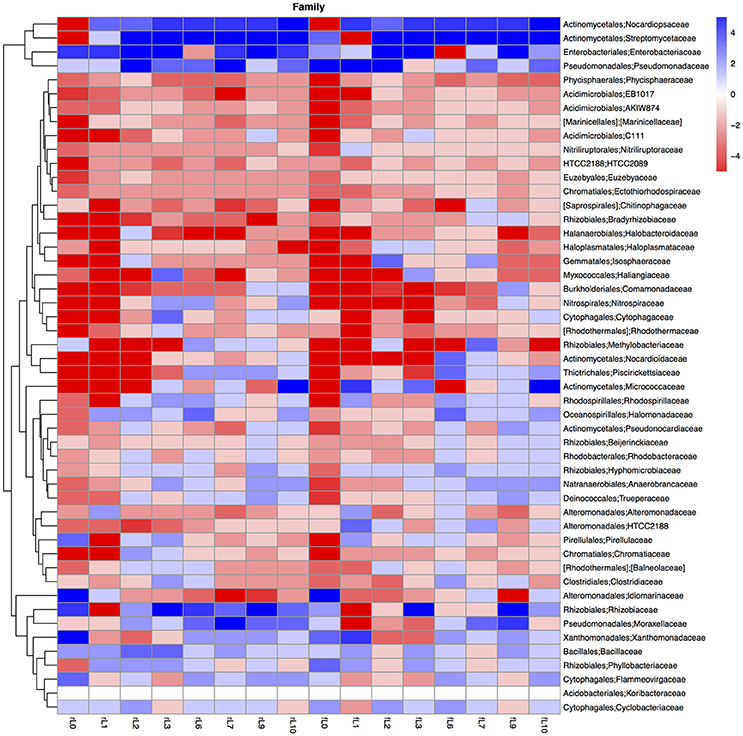
Figure 10. Heatmap of the ratio between the relative abundance of the different bacterial families in the unamended soil vs. the soil amended with maize plantlets or its neutral detergent fiber (NDF). Legends to the figure can be found in Figure 5.
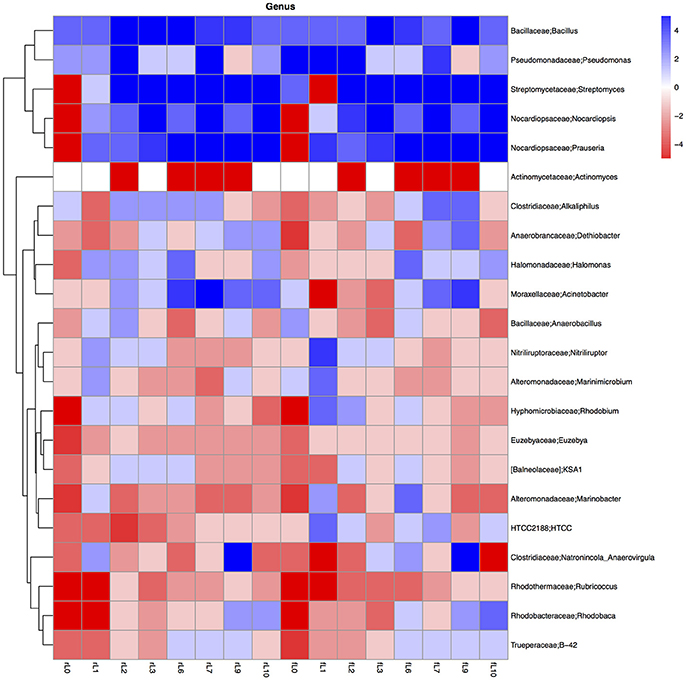
Figure 11. Heatmap of the ratio between the relative abundance of the different bacterial genera in the unamended soil vs. the soil amended with maize plantlets or its neutral detergent fiber (NDF). Legends to the figure can be found in Figure 5.
The ratio between the relative abundance of the bacterial groups in the NDF amended and the unamended soil with different salinity showed a similar pattern as that observed when maize plants were applied, except for phylotypes belonging to the Pseudomonadales (Figures 5, 8–11, Table S4). The relative abundance of the Pseudomonadales was highly different in the unamended soil, the NDF amended and maize amended soil with decreasing salt content. Phylotypes belonging to the Pseudomonadales participated in the degradation of the NDF in soil with EC ≥ 14.1 dS m−1, did not participate in soil with EC between ≤ 8.3 dS m−1 and ≥ 3.3 dS m−1 and participated again when EC ≤ 2.3 dS Phylotypes belonging to the Pseudomonadales did not participate in the degradation of the maize plants soil with EC ≥ 14.1 dS m−1, but participated when the EC dropped to ≤ 8.3 dS m−1. Interestingly, different patterns were also found when bacterial groups at a lower taxonomic level of Pseudomonadales were considered, e.g., Acinetobacter, other Pseudomonadaceae and Pseudomonas. For instance, the relative abundance of Pseudomonas increased sharply in the NDF amended soil with EC 157.8 dS m−1, but less so in the maize amended soil.
Discussion
Soil Characteristics
It is often difficult to define which factors control the bacterial community structure in an ecosystem as changes in one factor are often accompanied by changes in other soil factors. For instance, soil clay content is often related to sodium absorption ratio and pH (Oades, 1988; Nelson and Oades, 1998), organic matter dispersion and sodicity in soils (Norrström and Bergstedt, 2001; Rietz and Haynes, 2003; Wong et al., 2008). It was assumed that flooding the extremely saline Texcoco soil would decrease salt content, but not affect other soil characteristics so that the effect of EC on bacterial community could be determined. Monthly flooding of the Texcoco soil sharply decreased EC, but not pH, a factor known to affect the bacterial community structure even with small changes (Fuentes et al., 2006; Nacke et al., 2011; Kuramae et al., 2012; Lanzén et al., 2015).
In previous studies, it was found that Texcoco soil drained in the field for extended periods tended to have lower clay content than undrained soil. In this study clay content also gradually decreased due to flooding, so, it appears that clay particles were also washed out in the field with the drainage water. Clay content is known to affect the bacterial community structure in soil (Ling et al., 2014; Wei et al., 2014). Consequently, short-term changes in the bacterial community were controlled by changes in EC, but the gradual decrease in clay might also have affected the bacterial community structure.
Flooding the Soil and Its Effect on the Bacterial Community Structure
It is well-known that how bacteria respond to stress depends on the level of stress, the type of stress and environmental conditions. Salinity is a known stress factor, and its effects on microorganisms have been studied extensively in fresh water (Marine et al., 2013; Telesh et al., 2013; Dupont et al., 2014) and soils (Canfora et al., 2014; Van Horn et al., 2014; Santini et al., 2015).
Experiments usually involve the application of different concentrations of salt and determining how the bacterial community responds to these changes. These studies, however, do not take into account that the microorganisms have no time to adapt to changes in salt concentrations. Chowdhury et al. (2011) found that for different types of soils, 7 to 10 days was enough to stabilize soil respiration so conditioning the soil between each flooding for 1 month would allow bacteria to adapt to the reduced salt content.
During the aerobic incubation, the bacterial community structure in the unamended soil showed large fluctuations over time. These changes could be attributed to intrinsic variability of the soil samples (destructive soil sampling), manipulating the soil or changes due to the aerobic incubation. Changes in the bacterial community in soil flooded 10 times were small during the aerobic incubation. Changes in the unflooded soil were not due to the aerobic incubation but due to variations in the samples or/and manipulation of the soil. Manipulating the unflooded soil stressed the bacterial population more than in the 10-times flooded soil as the salt content was higher so the effect of these changes on the bacterial community were larger.
The bacterial community has been studied in gradients of saline soils and similar groups dominated in most studies, i.e., Actinobacteria, Firmicutes, and Proteobacteria (Hollister et al., 2010; Van Horn et al., 2014). Santini et al. (2015) found that the bacterial community in bauxite residue was similar to that found in some soda lakes and other alkaline saline wastes with dominance of Proteobacteria (40–80%), Firmicutes (2–22%), Actinobacteria (3–26%), and Bacteroidetes (2–10%). They are considered generalists and in this study phylotypes belonging to the Actinobacteria, Bacteroidetes, Firmicutes, and Proteobacteria were dominant in soil.
The decrease in salt content due to flooding had different effects on different bacterial groups. First, a decrease in salt content only had a limited effect on the relative abundance of bacterial groups and no clear pattern emerged, e.g., Bacteroidetes, Chloroflexi, Nitrospirae, and Proteobacteria. Barnard et al. (2013) considered Proteobacteria, Firmicutes, Gemmatimonadetes, Planctomycetes, and Chloroflexi to have a more stable and resistant life strategy as witnessed by a more stable ribosome content during a dry-down/wet-up experiment. The Chloroflexi and Proteobacteria were also resistant to changes in the salt content as found in this study. Second, the drop in salt content after the first flooding increased the relative abundance of bacterial groups but further decreases in salt content had only a limited effect on the relative abundance of the bacterial group, e.g., Actinobacteria, Armatimonadetes, and [Thermi]. Hollister et al. (2010) found that the relative abundance of Actinobacteria increased with decreasing water content and salinity. Third, the drop in salt content after the first flooding increased the relative abundance of bacterial groups and further decreases in salt content tended to further increase the relative abundance of the bacterial groups Gemmatimonadetes, Planctomycetes, TM7, and Verrucomicrobia. Canfora et al. (2014) also found a higher relative abundance of Gemmatimonadetes in the low salinity soil (EC 5.37 dS m−1). Fourth, decreasing the salt content decreased the relative abundance of bacterial groups, like Chlorobi and Firmicutes, which were thus well-adapted to a high soil salt content. Phylotypes belonging to the genus Bacillus (Firmicutes) are spore forming so easily survive in adverse conditions (Teixeira et al., 2010; van Djil and Hecker, 2013). Additionally, they are involved in the early decomposition of organic material that will be liberated when the soil was manipulated or/and microorganisms die due to the extreme adverse conditions (Cleveland et al., 2007; Hu B. et al., 2015; Hu P. et al., 2015).
Changes in the bacterial community structure could also be due to a loss of clay particles caused by flooding the soil. Clay content was the parameter that best demonstrated the bacterial community composition at OTU-97% level in the unamended soil. Sessitsch et al. (2002) found a relationship between soil particle size and the presence/absence of some bacterial groups, relating small size particles (clay) with a greater diversity.
Mineralization
It has been reported that microbial communities in saline soil are less efficient in using C sources than those in non-saline soils (Setia et al., 2010). Soil microorganisms respond differently to changes in the osmotic potential by either accumulating or excreting osmolites (Empadinhas and da Costa, 2008) or other metabolites (Kempf and Bremer, 1998; Oren, 2008), which are energy expensive processes (Oren, 2002). In this study, however, the emitted CO2 was the highest in the soil with the highest salt content. It can be assumed that flooding the soil drained easily decomposable organic material thereby reducing the CO2 emitted.
Yan and Marschner (2013) found an increase in C mineralization when pea straw was added to leached saline soils. In this study, the amount of CO2 emitted also increased when maize plants or its NDF fraction was applied to soil. The difference in CO2 emitted from the unamended soil and the organic material amended soil was the result of two processes, i.e., mineralization of the applied maize or its NDF fraction and a possible priming effect. A priming effect was measured in previous studies when maize plants were applied to the Texcoco soil and it decreased with decreased soil salt content (Conde et al., 2005). The increase in CO2 emitted after the application of organic material when the EC in soil decreased might be due to an increase in the amount of organic material mineralized and the decrease due to a decline in the priming effect. However, no 14C nor 13C labeled organic material were used in this study so a possible priming effect and changes in its magnitude due to changes in the soil salt content with flooding could not be determined nor could possible changes in the amount of applied organic material mineralized be measured.
Organic Material Addition, Flooding, and Their Effect on Bacterial Community Structure
Three different factors were investigated in this study. First, the effect of organic material application on the relative abundance of different bacterial groups was determined, second the effect of a reduction in soil salt content through flooding on the bacteria involved in organic material decomposition was studied, and third the effect of the composition of the organic material applied, i.e., maize plantlets vs. their NDF, on the bacterial community structure was investigated.
First, the application of organic material, i.e., maize plantlets and their NDF fraction, had a profound effect on the relative abundance of different bacterial groups. Organic material decomposition is primarily dominated by microorganisms that prefer easily decomposable organic matter and a higher nutrient content. These microorganisms are known as r-strategist or copiotrophs, while the k-strategists or oligotrophs prefer an environment with a low nutrient content and prevail when nutrients are depleted (Fierer et al., 2007; Trivedi et al., 2013). With a few exceptions, the number of bacterial groups of which the relative abundance increased when organic material was applied to soil was smaller than the number of bacterial groups of which the relative abundance decreased. As such, only a limited number of bacterial groups were favored by the application of organic material and metabolized maize plantlets and their NDF fraction. It has to be stipulated that only the relative abundance of the bacterial groups was determined. Application of organic material increases the soil microbial biomass so it can be assumed that absolute abundance of some bacterial groups was not affected although the relative abundance decreased. Additionally, fungi also participate in the degradation of organic material applied and the changes in their relative abundances were not determined.
Second, flooding the soil, which reduced the soil salt and clay content, affected the relative abundance of bacterial groups in soil amended with organic material compared to the unamended soil. The changes in soil characteristics as a result of flooding altered the metabolic capacity of some bacterial groups, i.e., they changed from a copiotrophic behavior to an oligotrophic behavior or vice versa, while the metabolic capacity of others was not affected. Classifying these metabolic changes with flooding allowed to define six categories of bacterial response to the application of maize plants or its NDF fraction. Considering the large amounts of bacterial groups retrieved from soil, e.g., 521 genera, only the most abundant ones will be discussed, although a complete list is given in Table 2.
The ratio of the relative abundance (Equation 1) for a first category of bacterial groups showed no clear pattern with flooding or the changes in their ratios were small. Hence, they participated sometimes or only to a limited degree in the degradation of the maize plants. They can be considered “undefined” as they sometimes behaved as oligotrophs and other times as copiotrophs with no clear effect of flooding. Phylotypes belonging to the Proteobacteria, the most dominant phylum, responded in this way. However, the different orders belonging to this phylum responded clearly, but differently to the application of organic material when the soil was flooded. Therefore, different patterns sometimes emerged within a bacterial group when different taxonomic levels were considered.
The relative abundance of a second category of bacterial groups mostly increased when maize plants were applied to the unamended soil (Table 2). Phylotypes belonging to the Firmicutes responded in this way. Phylotypes belonging to the Firmicutes are known copiotrophs (Chávez-Romero et al., 2016). For instance, Van Horn et al. (2014) applied organic material to soil from a natural salinity gradient (105 ± 4–4,800 ± 470 μS) at different water contents. They found an increase in the relative abundance of the Firmicutes in all soils. At a lower taxonomic level, members of the Bacillaceae (Bacillus) and Pseudomonadaceae (Pseudomonas) participated in the degradation of maize plants independent of the number of floodings. Both families have been described as decomposers of organic material in soil. Cahyani et al. (2003) found Bacillaceae throughout composting of rice straw and considered them to be saccharolytic bacteria. López-Lozano et al. (2013) found Bacillaceae and Pseudomonadaceae among other known opportunist heterotrophs. They found these groups to be initial colonizers after sterilizing and re-inoculating an arid soil and attributed this to the enzymatic capacity of members of this family to initiate the degradation of macromolecular carbon compounds. Yeasmin et al. (2015) found an increase in phylotypes belonging to the Bacillaceae in a paddy soil mixed with cow manure and rice straw. Members of Bacillus are known copiotrophs and always participated in the degradation of the maize plants independent of the soil salt content. Their metabolic capacity to participate in the degradation of maize plants was not hindered by a wide range of salt content and they were not outcompeted by other bacterial groups. The Cyclobacteriaceae were also favored mostly by the addition of maize plants or NDF. Members of Cyclobacterium are halophilic were recently isolated from a Korean fore shore soil (Irshad et al., 2014). Goldfarb et al. (2011) incubated soil in the presence of a range of C substrates varying in chemical recalcitrance and DNA analyzed by 16S rRNA PhyloChip. They found that the relative abundance of Enterobacteriales increased when labile organic substrates (i.e., glycine and sucrose) were applied to soil. In this study phylotypes belonging to the Enterobacteriales behaved mostly as copiotrophs and were not affected by the wide range of soil salt content.
A third category of bacterial groups, some well-known to participate in the degradation of maize plants, were inhibited metabolically or outcompeted by other bacterial groups in the soil with a high salt content, but their relative abundance increased compared to the unamended soil when the soil was flooded. They can be considered “saline adverse copiotrophs.” For instance, phylotypes belonging to the Actinomycetales, the most dominant order, were strongly inhibited when maize plants were applied to the unflooded soil (relative abundance of 1.1% in the unamended soil and 0.1% in the maize amended soil), but they participated in the decomposition of the maize plants and its degradation products when the soil was flooded once. Phylotypes belonging to Prauseria, the most dominant genus (Nocardiopsaceae, Actinomycetales), can also be considered saline adverse copiotrophs. Their metabolic capacity to participate in the degradation of maize plants was hindered by the high salt content and they were outcompeted by other bacterial groups (e.g., Bacillus). Phylotypes belonging to Halomonadaceae and Halomonas were also saline adverse copiotrophs. Members of Halomonas are halophilic (Meyer et al., 2015), but not extreme halophilic.
The fourth category with a limited number of bacterial groups participated in the degradation of maize plants in the unflooded soil, but they were not favored by application of organic material when the soil was flooded and can be considered “saline copiotrophs.” Phylotypes belonging to the Sphingobacteria responded in this way.
The relative abundance of a wide range of bacterial groups decreased when maize plants were applied to soil compared to the unamended soil. The decrease was similarly independent of flooding, i.e., a fifth category of bacterial groups or oligotrophs, or the decrease was more outspoken in the unflooded soil than in the flooded soil, i.e., a sixth category of bacterial groups or saline oligotrophs. Chloroflexi, Deltaproteobacteria, and Planctomycetes belonged to the fifth category of bacteria and their relative abundance always decreased in the same way in the maize plants amended soil compared to the unamended soil. The decrease in the relative abundance of Acidobacteria, Gemmatimonadetes, and Verrucomicrobia was more outspoken when maize plants were applied to the unflooded soil compared the unamended unflooded soil than when the soil was flooded. Most of these bacterial groups are known oligotrophs. For instance, Carbonetto et al. (2014) considered Chloroflexi and Verrucomicrobia oligotrophic. Siles et al. (2014) found a decrease in the relative abundance of Gemmatimonadaceae after the addition of dry olive residue to a Mediterranean soil in a 60 days incubation experiment.
de Lourdes Moreno et al. (2013) found that halophilic bacteria, such as Marinobacter lipolyticus produced different extracellular hydrolases (lipase, amylase, protease, xylanase, DNase, inulinase, pectinase, cellulose, and pulullanase), which gave them the capacity to degrade organic material in ecosystems with extreme salinity. In this study, however, phylotypes belonging to Marinobacter did participate in the degradation of maize plants when the soil was flooded once but not when the soil was not flooded. It might be that salinity was too high or characteristics other than salinity inhibited the metabolic capacity of Marinobacter in the Texcoco soil. Goldfarb et al. (2011) incubated soils in the presence of a range of C substrates varying in chemical recalcitrance and analyzed DNA by 16S rRNA PhyloChip. They found that the relative abundance of the Burkholderiales increased when labile or chemically recalcitrant substrate was applied to soil. Eichorst and Kuske (2012) conducted a SIP study in soil microcosms to identify cellulose responsive bacteria that actively use 13[C] cellulose in five edaphically different soils. They found members of the Burkholderiales in the five soils. In this study phylotypes belonging to the Burkholderiales behaved as saline oligotrophs. As mentioned before, it could be that the salt content of the soil was still too high and inhibited the metabolic capacity of the Burkholderiales or another soil factor inhibited them participating in the mineralization of the maize plants, e.g., the high pH.
Third, the application of maize plants mostly had a similar effect on the bacterial groups as the application of NDF. However, one bacterial group, the Pseudomonadales (e.g., Acinetobacter and Pseudomonas), was more favored by the application of NDF than when maize plants were applied to the soil with high salt contents. Pseudomonadales are versatile and phylotypes belonging to this bacterial group are known to have cellulolytic activity (Goldfarb et al., 2011). They metabolized (hemi)cellulose in the soils with high salt content.
Nearly 30% of the retrieved sequences in this experiment could not be assigned to a bacterial group. Santini et al. (2015) found the same for 90% of the bacterial and 70% of fungal OTUs in engineered haloalkaline soil, i.e., the salinity had increased due to anthropogenic activities. Although in their experiment some of those OTUs were shared with other natural and engineered environments. This gives us an idea of the existing gap in our knowledge and understanding of extreme environments as most of the microorganisms are yet to be described.
It has to be remembered that relative abundances of bacterial groups were determined and not absolute values. Application of easily decomposable organic material, such as the maize plants, will increase the soil microbial biomass so an increase in relative abundance of bacterial groups means that the absolute value also increased, but a decrease in relative abundance does not necessarily mean that the absolute abundance of the bacterial group decreased. This is most likely true when changes in the ratio between the relative abundance of a bacterial group in the unamended soil and the organic material amended soil were small. However, most of the changes reported in this study were large and the decrease or increase in the relative abundance due to the application of organic material were five times lower or higher than in the unamended soil.
Additionally, no unflooded soil was kept for 10 months to determine how the bacterial community structure might be altered in an unflooded soil over time. Soil organic matter will be mineralized so the C substrate available for the soil microorganisms will decrease and this might alter the bacterial community structure, but it was assumed that changes would be minimal.
Conclusion
Flooding the soil substantially reduced the salt content of the Texcoco soil and after ten monthly floodings, the electrolytic conductivity dropped to 1.7 dS m−1 from an initial 157.8 dS m−1. Flooding the soil also decreased the soil clay content, but the decrease was less accentuated than the decrease in EC, while the pH was not affected by flooding. The change in salt content was the factor that affected the relative abundance of bacterial populations the most. Decreasing the salt content altered the metabolic behavior of some bacterial groups, but not others, e.g., Nitriliruptor. Considering the ratio between the relative abundance of a bacterial group in the maize or neutral detergent fiber amended soil vs. the unamended soil allowed to distinguish different effects of salt content on the metabolic capacity of bacterial groups and to categorize them. Some bacterial groups, such as Bacillus and Gammaproteobacteria, did participate in the degradation of the organic material applied in a soil with a wide range of salinity, i.e., copiotrophs. Some copiotrophic bacteria were not favored by the application of organic material in soils with extreme salinity, e.g., Actinomycetales, Nocardiopsis, and Pauseria, while a limited number were not favored when the soil salt content decreased, e.g., Sphingobacteria. The first could be called “saline adverse copiotrophs” and the latter “saline copiotrophs”. Other bacterial groups, e.g., Deltaproteobacteria, were not favored by the application of the organic material and their relative abundance always decreased compared to the unamended soil, i.e., oligotrophs. The negative effect of application of organic material on some of these oligotrophs was extreme when the soil salt content was high, e.g., Acidobacteria, Chloroflexi, Gemmatimonadetes, and Verrucomicrobia, and the ratio of the relative abundance in the maize plants amended soil vs. the unamended soil was high. This negative effect decreased, however, with a decrease in soil salt content. They could be called “saline oligotrophs.” The effect of soil salt content on the participation or not of bacterial groups in the degradation of organic material was similar when maize plants or the neutral detergent fraction was applied to soil except for phylotypes belonging to the Pseudomonadales.
Author Contributions
AD-L: Participated in the experiment, writing of manuscript, and data analysis. LD-B: Did the experiment. CD-M: Participated in the Experiment. YN-N: Contributed to the Experimental design, data analysis, writing of manuscript. ML-G: Contributed to the Experimental design, writing of manuscript. LD: Conceived the experiment, experimental design, writing of manuscript, data analysis.
Funding
The research was funded by Instituto de Ciencia y Tecnología del Distrito Federal (ICTyDF, México), Apoyo Especial para Fortalecimiento de Doctorado PNPC 2013, 2014 and project Infraestructura 205945 from Consejo Nacional de Ciencia y Tecnología (CONACyT, Mexico). This research was also funded by Centro de Investigación y de estudios avanzados del Instituto Politécnico Nacional (Cinvestav). AD-L, CD-M, and LD-B received grant-aided support from CONACyT, while YN-N from ABACUS-CONACyT.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank ABACUS (CONACyT) for the use of the computer to denoise the sequences and Shona Prince Laurette for the English corrections.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2018.00051/full#supplementary-material
References
Acinas, S., Klepac-Ceraj, V., Hunt, D., Pharino, C., Ceraj, I., Distel, D., et al. (2004). Fine-scale phylogenetic architecture of a complex bacterial community. Nature 430, 551–554. doi: 10.1038/nature02649
Barnard, R., Osborne, C., and Firestone, M. (2013). Responses of soil bacterial and fungal communities to extreme desiccation and rewetting. ISME J. 7, 2229–2241. doi: 10.1038/ismej.2013.104
Baumann, K., and Marschner, P. (2013). Effects of salinity on microbial tolerance to drying and rewetting. Biogeochemistry 112, 71–80. doi: 10.1007/s10533-011-9672-1
Bellini, G., Sumner, M., Radcliffe, D., and Qafoku, N. (1996). Anion transport trough columns of highly weathered acid soil: adsorption and retardation. Soil Sci. Soc. Am. J. 60, 132–137. doi: 10.2136/sssaj1996.03615995006000010021x
Beltrán-Hernández, R., Luna-Guido, M., and Dendooven, L. (2007). Emission of carbon dioxide and dynamics of inorganic N in a gradient of alkaline saline soils of the former lake Texcoco. Appl. Soil Ecol. 35, 390–403. doi: 10.1016/j.apsoil.2006.07.005
Cahyani, V., Matsuya, K., Asakawa, S., and Kimura, M. (2003). Succession and phylogenetic composition of bacterial communities responsible for the compositing process of rice straw estimated by PCRD-GGE analysis. J. Soil Sci. Plant Nutr. 49, 619–630. doi: 10.1080/00380768.2003.10410052
Canfora, L., Bacci, G., Pinzari, F., Papa, G. L., Dazzi, C., and Benedetti, A. (2014). Salinity and bacterial diversity: to what extent does the concentration of salt affect the bacterial community in a saline soil? PLoS ONE 9:e106662. doi: 10.1371/journal.pone.0106662
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010a). QIIME allows analysis of high–throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Caporaso, J., Bittinger, K., Bushman, F., DeSantis, T., Andersen, G., and Knight, R. (2010b). PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26, 266–267. doi: 10.1093/bioinformatics/btp636
Carbonetto, B., Rascovan, N., Álvarez, R., Mentaberry, A., and Vázquez, M. (2014). Structure, composition and metagenomic profile of soil microbiomes associated to agricultural land use and tillage systems in Argentine Pampas. PloS ONE 9:e99949. doi: 10.1371/journal.pone.0099949
Carrigg, C., Rice, O., Kavanagh, S., Collins, G., and O'Flaherty, V. (2007). DNA extraction method affects microbial community profiles from soils and sediment. Appl. Microbiol. Biotechnol. 77, 955–964. doi: 10.1007/s00253-007-1219-y
Chávez-Romero, Y., Navarro-Noya, Y., Reynoso-Martínez, S., Sarria-Guzmán, Y., Govaerts, B., Verhulst, N., et al. (2016). 16S metagenomics reveals changes in the soil bacterial community driven by soil organic C, N-fertilizer and tillage-crop residue management. Soil Tillage Res. 159, 1–8. doi: 10.1016/j.still.2016.01.007
Chowdhury, N., Marschner, P., and Burns, R. (2011). Response of microbial activity and community structure to decreasing soil osmotic and matric potential. Plant Soil 344, 241–254. doi: 10.1007/s11104-011-0743-9
Clarke, K., and Ainsworth, M. (1993). A method of linking multivariate community structure to environmental variables. Mar. Ecol. Prog. Ser. 92, 205–219. doi: 10.3354/meps092205
Cleveland, C., Nemergut, D., Schmidt, S., and Townsend, A. (2007). Increases in soil respiration following labile carbon additions linked to rapid shifts in soil microbial community composition. Biogeochemistry 82, 229–240. doi: 10.1007/s10533-006-9065-z
Coleman, D. C., and Wall, D. (2014). “Soil fauna: occurrence, biodiversity and roles in ecosystem function,” in Soil Microbiology, Ecology and Biochemistry, 4th Edn, ed E. A. Paul (San Diego, CA: Academic Press), 111–150.
Conde, E., Cardenas, M., Ponce-Mendoza, A., Luna-Guido, M., Cruz-Mondragón, C., and Dendooven, L. (2005). The impacts of inorganic nitrogen application on mineralization of 14 C-labelled maize and glucose, and on priming effect in saline alkaline soil. Soil Biol. Biochem. 37, 681–691. doi: 10.1016/j.soilbio.2004.08.026
de León-Lorenzana, A. S., Delgado-Balbuena, L., Domínguez-Mendoza, C., Navarro-Noya, Y. E., Luna-Guido, M., and Dendooven, L. (2017). Reducing salinity by flooding an extremely alkaline and saline soil changes the bacterial community but its effect on the archaeal community is limited. Front. Microbiol. 8:466. doi: 10.3389/fmicb.2017.00466
de Lourdes Moreno, M., Pérez, D., García, M., and Mellado, E. (2013). Halophilic bacteria as a source of novel hydrolytic enzymes. Life 3, 38–51. doi: 10.3390/life3010038
Dendooven, L., Ramirez-Fuentes, E., Alcantara-Hernandez, R., Valenzuela-Encinas, C., Sanchez-Lopez, K., Luna-Guido, M., et al. (2015). Dynamics of C-14-labelled glucose and NH4+ in a regularly flooded extremely alkaline saline soil. Pedosphere 25, 230–239. doi: 10.1016/S1002-0160(15)60008-X
Dungait, J., Hopkins, D., Gregory, A., and Whitmore, A. (2012). Soil organic matter turnover is governed by accessibility not recalcitrance. Glob. Change Biol. 18, 1781–1796. doi: 10.1111/j.1365-2486.2012.02665.x
Dupont, C. L., Larsson, J., Yooseph, S., Ininbergs, K., Goll, J., Asplund-Samuelsson, J., et al. (2014). Functional tradeoffs underpin salinity-driven divergence in microbial community composition. PLoS ONE 9:e89549. doi: 10.1371/journal.pone.0089549
Edgar, R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. doi: 10.1093/bioinformatics/btq461
Eichorst, S., and Kuske, C. (2012). Identification of cellulose-responsive bacterial and fungal communities in geographically and edaphically different soils by using stable isotope probing. Appl. Environ. Microbiol. 78. 2316–2327. doi: 10.1128/AEM.07313-11
Empadinhas, N., and da Costa, M. (2008). Osmoadaptation mechanisms in prokaryotes: distribution of compatible solutes. Int. Microbiol. 11, 151–161. doi: 10.2436/20.1501.01.55
Fierer, N., Bradford, M., and Jackson, R. (2007). Toward an ecological classification of soil bacteria. Ecology 88, 1354–1364. doi: 10.1890/05-1839
Fuentes, J., Bezdicek, D., Flury, M., Albrecht, S., and Smith, J. (2006). Microbial activity affected by lime in a long-term no-till soil. Soil Tillage Res. 88, 123–131. doi: 10.1016/j.still.2005.05.001
Gee, G. W., and Bauder, J. W. (1986). “Particle size analysis,” in Methods of Soil Analysis, Part 1. Physical and Mineralogical Methods, ed A. Klute (Madison, WI: American Society of Agronomy-Soil Science Society of America), 383–411.
Gennari, M., Abbate, C., La Porta, V., and Baglieri, A. (2007). Microbial response to Na2SO4 additions in a volcanic soil. Arid Land Res. Manag. 21, 211–227. doi: 10.1080/15324980701428732
Goldfarb, K., Karaoz, U., Hanson, C., Santee, C., Bradford, M., Treseder, K., et al. (2011). Differential growth responses of soil bacterial taxa to carbon substrates of varying chemical recalcitrance. Front. Microbiol. 2:94. doi: 10.3389/fmicb.2011.00094
Haas, B., Gevers, D., Earl, A., Feldgarden, M., Ward, D., Giannoukos, G., et al. (2011). Chimeric 16S rRNA sequence formation and detection in Sanger and 454–pyrosequenced PCR amplicons. Genome Res. 21, 494–504. doi: 10.1101/gr.112730.110
Hoffman, C., and Winston, F. (1987). A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene 57, 267–272. doi: 10.1016/0378-1119(87)90131-4
Hollister, E., Engledow, A., Hammett, A., Provin, T., Wilkinson, H., and Gentry, T. (2010). Shifts in microbial community structure along an ecological gradient of hypersaline soils and sediments. ISME J. 4, 829–838. doi: 10.1038/ismej.2010.3
Hu, B., Yang, Q., Cai, M., Tang, Y., Zhao, G., and Wu, X. (2015). Negadavirga shengliensis gen. nov., sp. nov., a novel member of the family Cyclobacteriaceae isolated from oil-contaminated saline soil. Antonie Van Leeuwenhoek 107, 663–673. doi: 10.1007/s10482-014-0361-7
Hu, P., Hollister, E., Somenahally, A., Hons, F., and Gentry, T. (2015). Soil bacterial and fungal communities respond differently to various isothiocyanates added for biofumigation. Front. Microbiol. 5:729. doi: 10.3389/fmicb.2014.00729
Irshad, A., Ahmad, I., and Kim, S. (2014). Culturable diversity of halophilic bacteria in foreshore soils. Braz. J. Microbiol. 45, 563–572. doi: 10.1590/S1517-83822014005000050
Jenkinson, D., and Powlson, D. (1976). The effects of biocidal treatments on metabolism in soil-V. A method for measuring soil biomass. Soil Biol. Biochem. 8, 209–213. doi: 10.1016/0038-0717(76)90005-5
Kempf, B., and Bremer, E. (1998). Uptake and synthesis of compatible solutes as microbial stress responses to high-osmolality environments. Arch. Microbiol. 170, 319–330. doi: 10.1007/s002030050649
Kuramae, E., Yergeau, E., Wong, L., Pijl, A., van Veen, J., and Kowalchuk, G. (2012). Soil characteristics more strongly influence soil bacterial communities than land-use type. FEMS Microbiol. Ecol. 79, 12–24. doi: 10.1111/j.1574-6941.2011.01192.x
Lanzén, A., Epelde, L., Garbisu, C., Anza, M., Martín-Sánchez, I., Blanco, F., et al. (2015). The Community structures of prokaryotes and fungi in mountain pasture soils are highly correlated and primarily influenced by pH. Front. Microbiol. 6:1321. doi: 10.3389/fmicb.2015.01321
Ling, N., Sun, Y., Ma, J., Guo, J., Zhu, P., Peng, C., et al. (2014). Response of the bacterial diversity and soil enzyme activity in particle-size fractions of Mollisol after different fertilization in a long-term experiment. Biol. Fertil. Soils 50, 901–911. doi: 10.1007/s00374-014-0911-1
López-Lozano, N., Heidelberg, K., Nelson, W., García-Oliva, F., Eguiarte, L., and Souza, V. (2013). Microbial secondary succession in soil microcosms of a desert oasis in the Cuatro Cienegas Basin, Mexico. PeerJ 1:e47. doi: 10.7717/peerj.47
Luna-Guido, M., Beltrán-Hernández, R., Solis-Ceballos, N., Hernández-Chavez, N., Mercado-García, F., Olalde-Portugal, V., et al. (2000). Chemical and biological characteristics of alkaline saline soils from the former Lake Texcoco as affected by artificial drainage. Biol. Fertil. Soils 32, 102–108. doi: 10.1007/s003740000223
Mamilov, A., Dilly, O. M., Mamilov, S., and Inubushi, K. (2004). Microbial eco-physiology of degrading aral sea wetlands: consequences for C-cycling. Soil Sci. Plant Nutr. 50, 839–842. doi: 10.1080/00380768.2004.10408544
Marine, C., Thierry, B., Olivier, P., Emma, R., Corine, B., Martin, A., et al. (2013). Freshwater prokaryote and virus communities can adapt to a controlled increase in salinity through changes in their structure and interactions. Estuar. Coast. Shelf Sci. 133, 58–66. doi: 10.1016/j.ecss.2013.08.013
Meyer, J., Dillard, B., Rodgers, J., Ritchie, K., Paul, V., and Teplitski, M. (2015). Draft genome sequence of Halomonas meridiana R1t3 isolated from the surface microbiota of the Caribbean Elkhorn coral Acropora palmata. Stand. Genomic Sci. 10, 75. doi: 10.1186/s40793-015-0069-y
Nacke, H., Thürmer, A., Wollherr, A., Will, C., Hodac, L., Herold, N., et al. (2011). Pyrosequencing-based assessment of bacterial community structure along different management types in German forest and grassland soils. PLoS ONE 6:e17000. doi: 10.1371/journal.pone.0017000
Navarro-Noya, Y., Gómez-Acata, S., Montoya-Ciriaco, N., Rojas-Valdez, A., Suárez-Arriaga, M., Valenzuela-Encinas, C., et al. (2013). Soil bacterial community structures in soil with different tillage and residue management: a metagenetic analysis. Soil Biol. Biochem. 65, 86–95. doi: 10.1016/j.soilbio.2013.05.009
Nelson, P., and Oades, J. (1998). “Organic matter, sodicity and soil structure,” in: Sodic Soils: Distribution, Properties, Management and Environmental Consequences. Topics in Sustainable Agronomy, eds M. E. Sumner and R. Naidu (New York, NY: Oxford University Press), 51–75.
Nilakanta, H., Drews, K. L., Firrell, S., Foulkes, M. A., and Jablonski, K. A. (2014). A review of software for analyzing molecular sequences. BMC Res. Notes 7:830. doi: 10.1186/1756-0500-7-830
Norrström, A., and Bergstedt, E. (2001). The impact of road de-icing salts (NaCl) on colloid dispersion and base cation pools in roadside soils. Water Air Soil Pollut. 127, 281–299. doi: 10.1023/A:1005221314856
Oades, J. (1988). The retention of organic matter in soils. Biogeochemistry 5, 35–70. doi: 10.1007/BF02180317
Oren, A. (2002). Diversity of halophilic microorganisms: environments, phylogeny, physiology, and applications. J. Ind. Microbiol. Biotechnol. 28, 56–63. doi: 10.1038/sj/jim/7000176
Oren, A. (2008). Microbial life at high salt concentrations: phylogenetic and metabolic diversity. Saline Syst. 4, 1. doi: 10.1186/1746-1448-4-2
Pathak, H., and Rao, D. (1998). Carbon and nitrogen mineralization from added organic matter in saline and alkaline soils. Soil Biol. Biochem. 30, 695–702. doi: 10.1016/S0038-0717(97)00208-3
Powlson, D., Gregory, P. J., Whalley, W. R., Quinton, J. N., Hopkins, D. W., Whitmore, A. P., et al. (2011). Soil management in relation to sustainable agriculture and ecosystem services. Food Policy 36, S72–S87. doi: 10.1016/j.foodpol.2010.11.025
Price, M., Dehal, P., and Arkin, A. (2009). FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 26, 1641–1650. doi: 10.1093/molbev/msp077
Ramírez-Villanueva, D., Bello-López, J., Navarro-Noya, Y., Luna-Guido, M., Verhulst, N., Govaerts, B., et al. (2015). Bacterial community structure in maize residue amended soil with contrasting management practices. Appl. Soil Ecol. 90, 49–59. doi: 10.1016/j.apsoil.2015.01.010
Rath, K., and Rousk, J. (2015). Salt effects on the soil microbial decomposer community and their role in organic carbon cycling: a review. Soil Biol. Biochem. 81, 108–123. doi: 10.1016/j.soilbio.2014.11.001
R Development Core Team (2008). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing.
Reeder, J., and Knight, R. (2010). Rapid denoising of pyrosequencing amplicon data: exploiting the rank-abundance distribution. Nat. Methods 7:668. doi: 10.1038/nmeth0910-668b
Rietz, D., and Haynes, R. (2003). Effects of irrigation-induced salinity and sodicity on soil microbial activity. Soil Biol. Biochem. 35, 845–854. doi: 10.1016/S0038-0717(03)00125-1
Ruíz-Valdiviezo, V., Luna-Guido, M., Galzy, A., Gutierrez-Miceli, F., and Dendooven, L. (2010). Greenhouse gas emissions and C and N mineralization in soils of Chiapas (Mexico) amended with leaves of Jatropha curcas L. Appl. Soil Ecol. 46, 17–25. doi: 10.1016/j.apsoil.2010.06.002
Sambrook, J., and Russell, D. W. (2001). Molecular Cloning: A Laboratory Manual. 3rd Edn. New York, NY: Cold Spring Harbor Laboratory Press.
Santini, T., Warren, L., and Kendra, K. (2015). Microbial diversity in engineered haloalkaline environments shaped by shared geochemical drivers observed in natural analogues. Appl. Environ. Microbiol. 81, 5026–5036. doi: 10.1128/AEM.01238-15
Sardinha, M., Müller, T., Schmeisky, H., and Joergensen, R. (2003). Microbial performance in soils along a salinity gradient under acidic conditions. Appl. Soil Ecol. 23, 237–244. doi: 10.1016/S0929-1393(03)00027-1
Sarig, S., and Steinberger, Y. (1994). Microbial biomass response to seasonal fluctuations in soil salinity under canopy of desert halophytes. Soil Biol Biochem,. 26, 1405–1408.
SAS Institute (1989). Statistic Guide for Personal Computers. Version 6.0. (ed). Cary, NC: SAS Institute.
Schimel, J., and Schaeffer, S. (2012). Microbial control over carbon cycling in soil. Front. Microbiol. 3:348. doi: 10.3389/fmicb.2012.00348
Sessitsch, A., Reiter, B., Pfeifer, U., and Wilhelm, E. (2002). Cultivation-independent population analysis of bacterial endophytes in three potato varieties based on eubacterial and Actinomycetes-specific PCR of 16S rRNA genes. FEMS Microbiol. Ecol. 39, 23–32. doi: 10.1111/j.1574-6941.2002.tb00903.x
Setia, R., and Marschner, P. (2013). Carbon mineralization in saline soils as affected by residue composition and water potential. Biol. Fertil. of Soils. 49, 71–77. doi: 10.1007/s00374-012-0698-x
Setia, R., Marschner, P., Baldock, J., and Chittleborough, D. (2010). Is CO2 evolution in saline soils affected by an osmotic effect and calcium carbonate? Biol. Fertil. Soils 46, 781–792. doi: 10.1007/s00374-010-0479-3
Siegwald, L., Touzet, H., Lemoine, Y., Hot, D., Audebert, C., and Caboche, S. (2017). Assessment of common and emerging bioinformatics pipelines for targeted metagenomics. PLoS ONE 12:e0169563. doi: 10.1371/journal.pone.0169563
Siles, J., Rachid, C., Sampedro, I., García-Romera, I., and Tiedje, J. (2014). Microbial diversity of a Mediterranean soil and its changes after biotransformed dry olive residue amendment. PLoS ONE 9:e103035. doi: 10.1371/journal.pone.0103035
Steiner, A. A. (1961). A universal method for preparing nutrient solutions of a certain desired composition. Plant Soil 15, 134–154. doi: 10.1007/BF01347224
Stockmann, U., Adams, M. A., Crawford, J. W., Field, D. J., Henakaarchchi, N., et al. (2013). The knowns, known unknowns and unknowns of sequestration of soil organic carbon. Agric. Ecosyst. Environ. 164, 80–99. doi: 10.1016/j.agee.2012.10.001
Strickland, M. S., Lauber, C., Fierer, N., and Bradford, M. A. (2009). Testing the functional significance of microbial community composition. Ecology 90, 441–451. doi: 10.1890/08-0296.1
Teixeira, L., Peixoto, R., Cury, J., Sul, W., Pellizari, V., Tiedje, J., et al. (2010). Bacterial diversity in rhizosphere soil from Antarctic vascular plants of Admiralty Bay, maritime Antarctica. ISME J. 4, 989–1001. doi: 10.1038/ismej.2010.35
Telesh, I., Schubert, H., and Skarlato, S. (2013). Life in the salinity gradient: discovering mechanisms behind a new biodiversity pattern. Estuar. Coast. Shelf Sci. 135, 317–327. doi: 10.1016/j.ecss.2013.10.013
Tripathi, S., Kumari, S., Chakraborty, A., Gupta, A., Chakraborty, K., and Bandyapadhyay, B. (2006). Microbial biomass and its activities in salt-affected coastal soils. Biol. Fertil. Soils 42, 273–277. doi: 10.1007/s00374-005-0037-6
Trivedi, P., Anderson, I., and Singh, B. (2013). Microbial modulators of soil carbon storage : integrating genomic and metabolic knowledge for global prediction. Trends Microbiol. 21:641–651. doi: 10.1016/j.tim.2013.09.005
Valenzuela-Encinas, C., Neria-González, I., Alcántara-Hernández, R., Enriques-Aragón, J., Hernandez-Rodriguez, C., Dendooven, L., et al. (2008). Phylogenetic analysis of archaeal community in an alkaline-saline soil of the former lake Texcoco (Mexico). Extremophiles 12, 247–254. doi: 10.1007/s00792-007-0121-y
Valenzuela-Encinas, C., Neria-González, I., Alcántara-Hernández, R. J., Estrada-Alvarado, I., Zavala-Díaz de la Serna, F. J., Dendooven, L., et al. (2009). Changes in the bacterial populations of the highly alkaline saline soil of the former lake Texcoco (Mexico) following flooding. Extremophiles 13, 609–621. doi: 10.1007/s00792-009-0244-4
van Djil, J., and Hecker, M. (2013). Bacillus subtilis: from soil bacterium to super-secreting cell factory. Microb. Cell Fact. 12:1. doi: 10.1186/1475-2859-12-3
Van Horn, D., Jordan, G., Okie, H., Buelow, N., Michael, N., Gooseff, J., et al. (2014). Soil microbial responses to increased moisture and organic resources along a salinity gradient in a Polar Desert. Appl. Environ. Microbiol. 80, 3034–3043. doi: 10.1128/AEM.03414-13
Van Soest, P. J. (1963). Use of detergents in the analysis of fibrous feeds. 2. A rapid method for the determination of fiber and lignin. J. AOAC Int. 46, 829–835.
Van Soest, P., and Wine, R. (1967). Use of detergents in the analysis of fibrous feeds. IV. Determination of plant cell-wall constituents. J. Assoc. Off. Anal. Chem. 50, 50–55.
Wang, Q., Garrity, G., Tiedje, J., and Cole, J. (2007). Naïve bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. doi: 10.1128/AEM.00062-07
Wei, X., Shao, M., Gale, W., and Li, L. (2014). Global pattern of soil carbon losses due to the conversion of forests to agricultural land. Sci. Rep. 4:4062. doi: 10.1038/srep04062
Wichern, J., Wichern, F., and Joergensen, R. G. (2006). Impact of salinity on soil microbial communities and the decomposition of maize in acidic soils. Geoderma 137, 100–108. doi: 10.1016/j.geoderma.2006.08.001
Wieder, W., Bonan, G., and Allison, S. (2013). Global soil carbon projections are improved by modelling microbial processes. Nat. Clim. Chang. 3, 909–912. doi: 10.1038/nclimate1951
Wong, V., Dalal, R., and Greene, R. (2008). Salinity and sodicity effects on respiration and microbial biomass of soil. Biol. Fertil. Soils 44, 943–953. doi: 10.1007/s00374-008-0279-1
Wong, V., Dalal, R., and Greene, R. (2009). Carbon dynamics of sodic and saline soils following gypsum and organic material additions: a laboratory incubation. Appl. Soil Ecol. 41, 29–40. doi: 10.1016/j.apsoil.2008.08.006
Wong, V., Greene, R., Dalal, R., and Murphy, B. (2010). Soil carbon dynamics in saline and sodic soils: a review. Soil Use Manage. 26, 2–11. doi: 10.1111/j.1475-2743.2009.00251.x
Yan, N., and Marschner, P. (2013). Microbial activity and biomass recover rapidly after leaching of saline soils. Biol. Fertil. Soils 49, 367–371. doi: 10.1007/s00374-012-0733-y
Yeasmin, S., Kim, C., Islam, S., and Lee, J. (2015). Population dynamics of cellulolytic bacteria depend on the richness of cellulosic materials in the habitat. Microbiology 84, 278–289. doi: 10.1134/S0026261715020186
Keywords: alkalinity, carbon mineralization, regular flooding, soil characteristics, copiotroph, oligotroph
Citation: De León-Lorenzana AS, Delgado-Balbuena L, Domínguez-Mendoza CA, Navarro-Noya YE, Luna-Guido M and Dendooven L (2018) Soil Salinity Controls Relative Abundance of Specific Bacterial Groups Involved in the Decomposition of Maize Plant Residues. Front. Ecol. Evol. 6:51. doi: 10.3389/fevo.2018.00051
Received: 18 April 2017; Accepted: 10 April 2018;
Published: 14 May 2018.
Edited by:
Ludmilla Aristilde, Cornell University, United StatesReviewed by:
Fernando José Cebola Lidon, Faculdade de Ciências e Tecnologia da Universidade Nova de Lisboa, PortugalSuprasanna Penna, Bhabha Atomic Research Centre, India
Copyright © 2018 De León-Lorenzana, Delgado-Balbuena, Domínguez-Mendoza, Navarro-Noya, Luna-Guido and Dendooven. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Luc Dendooven, dendooven@me.com
 Arit S. De León-Lorenzana1
Arit S. De León-Lorenzana1  Cristina A. Domínguez-Mendoza
Cristina A. Domínguez-Mendoza Yendi E. Navarro-Noya
Yendi E. Navarro-Noya Luc Dendooven
Luc Dendooven