Synergies Between Division of Labor and Gut Microbiomes of Social Insects
- 1Section for Ecology and Evolution, Department of Biology, University of Copenhagen, Copenhagen, Denmark
- 2Centre for Biodiversity and Environment Research, University College London, London, United Kingdom
- 3Department of Fundamental Microbiology, University of Lausanne, Lausanne, Switzerland
- 4Université Clermont Auvergne, INRA, UMR 454 MEDIS, 63000, Clermont-Ferrand, France
- 5UMR UPEC, IRD 242, CNRS 7618, UPMC 113, INRA 1392, PARIS 7 113, Institut d'Ecologie et des Sciences de l'Environnement de Paris, Créteil, France
- 6Institut National de la Santé et de la Recherche Médicale, Paris, France
Social insects maximize resource acquisition and allocation through division of labor and associations with microbial symbionts. Colonies divide labor among castes and subcastes, where the plasticity of caste roles decreases in clades with higher social grades. Recent studies indicate that specific castes may also foster distinct gut microbiomes, suggesting synergies between division of labor and symbiosis. The social organization of a colony potentially partitions evolutionary persistent microbial partners to optimize symbioses and complement division of labor. However, research in this area has received limited attention. To elucidate if a structured microbiota is adaptive, we present three testable predictions to address consistent community structure, beneficial functions, and selection for microbiota that support caste roles. First, we posit that social insect groups spanning lower to higher social grades exhibit increasingly distinct caste microbiomes, suggesting that structured microbiomes may have evolved in parallel to social complexity. Second, we contend that the development of these microbiomes during colony maturation may clarify the extent to which they support division of labor. Third, we predict that mature social insect colonies with the most extreme division of labor demonstrate the strongest distinctions between caste microbiomes, carrying the greatest promise of insight into microbiome composition and function. Ultimately, we hypothesize that caste-specific microbiomes may enhance symbiotic benefits and the efficiency of division of labor, consequently maximizing fitness.
Introduction
Organisms are selected to optimize resource use through their own actions and interactions, in turn maximizing reproductive success. Ants, termites, and the social bees and wasps compose more than half of the biomass of land-dwelling insects (Hölldobler and Wilson, 2009) and have comprehensive ecological impacts as pollinators, predators, herbivores, and decomposers (Bignell and Eggleton, 2000; Richter, 2000; Aizen et al., 2008; Del Toro et al., 2012). Their success has, in part, been attributed to their division of labor (Oster and Wilson, 1978), where castes within the colony optimally divide tasks and thus improve the efficiency by which resources are amassed, distributed, and utilized (Anderson and Ratnieks, 1999; Duarte et al., 2011).
Microbial gut symbionts augment social insect metabolism and defense, further allowing the colonies to monopolize various resources. Termites digest recalcitrant plant substrates with obligate gut symbionts that degrade lignocellulose and upgrade dietary nitrogen (Cleveland, 1923; Brune, 2014). Social corbiculate bees host bacterial communities that aid in nutrient acquisition from pollen (Kwong and Moran, 2016; Kesnerova et al., 2017; Zheng et al., 2017). Herbivorous ants maintain gut bacteria that offset host dietary and metabolic limitations through amino acid supplementation, nitrogen recycling, and catabolism of glucose and citrate (Russell et al., 2009; Hu et al., 2018; Sapountzis et al., 2018). Gut microbes also contribute to disease defense in bees and termites by imparting pathogen colonization resistance (Koch and Schmid-Hempel, 2011; Peterson and Scharf, 2016; Raymann et al., 2017; Inagaki and Matsuura, 2018), further enabling host persistence and resource acquisition across environments.
The colony gut microbiota may be organized and optimized to complement division of labor, with castes partitioning the gut microbiota to support specialized roles. This could potentially enhance symbiont productivity to meet host needs and increase the efficiency of division of labor. A partitioned microbiota may incur a selective benefit if it improves resource acquisition and allocation. To begin to assess this hypothesis, we review division of labor in the social insects, known gut microbiota compositional differences between castes and subcastes, and the role of social interactions in influencing persistent gut microbiota. We then discuss three future avenues of research that may allow insights into putative interfaces between social organization and microbiomes: (1) structure and function of colony microbiomes across social insects that vary in social complexity, (2) development of caste microbiomes during colony maturation, and (3) caste-specific gut microbiomes within colonies with the most extreme division of labor.
Division of Labor is a Cornerstone in Social Insect Biology
Colony-level social complexity of social insect species is dictated by division of labor between worker and reproductive castes. Workers perform non-reproductive tasks such as brood care, defense, and foraging while the reproductive caste comprises one or a few individuals that secure colony fecundity. Variation in the degree of this division of labor can be interpreted along an evolutionary gradient of increasing social complexity and decreasing individual-level reproductive plasticity (Taylor et al., 2019) (Figure 1). In the most derived social insects, workers are morphologically precluded from attaining a reproductive role, resulting in strong suppression of inter-caste reproductive conflict (Boomsma and Gawne, 2018).
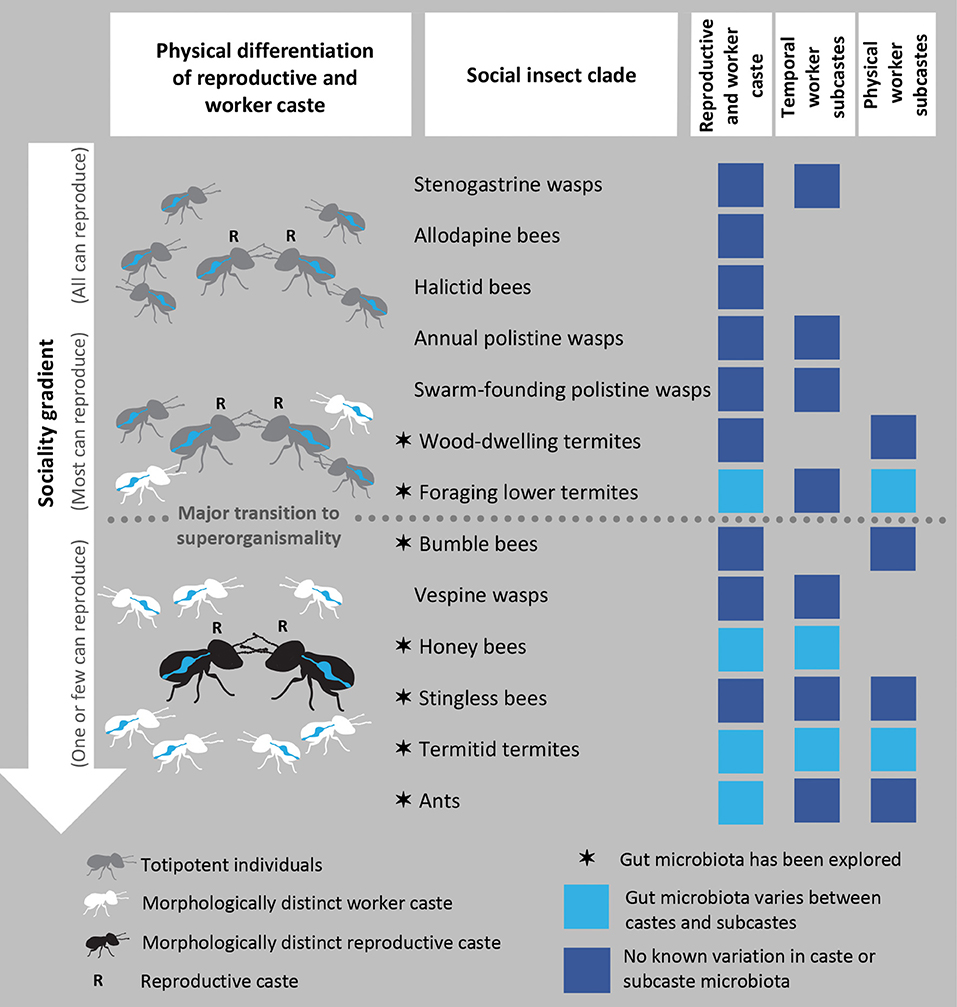
Figure 1. The sociality gradient and gut microbial symbionts of social insects. The sociality gradient is illustrated by the greyscale changes in the insects, indicative of reductions in individual reproductive plasticity. The gradient begins with colonies where totipotent workers help reproductives through adulthood, to which subsocial family life is a critical evolutionary precursor and culminates in superorganismal colonies with morphologically distinct castes. The position of clades along the gradient is adapted from Boomsma and Gawne (2018). Clades for which the gut microbiota has been explored are marked with a star, while evidence or lack thereof on distinct (sub)caste microbiomes is represented by light and dark blue squares, respectively. The first column of boxes illustrates that all clades divide reproductive labor between the reproductive and worker caste. The second and third columns signify if members of the clade have specialized division of non-reproductive labor among temporal and physical worker subcastes (temporal: bees, Seeley, 1982; Wille, 1983; termites, Watson and Sewell, 1985; ants, Hölldobler and Wilson, 1990; physical: bees, Goulson et al., 2002; Spaethe and Weidenmüller, 2002; Evans, 2006; wasps, Jandt and Toth, 2015; Li et al., 2015; Du et al., 2017; Grüter et al., 2017; termites, Korb and Thorne, 2017).
Morphological reproductive-worker differentiation and repression of conflict between interdependent castes represent a major evolutionary transition to higher complexity (Maynard Smith and Szathmáry, 1995; West et al., 2015). This transition aligns the fitness interests of all colony members, with selection acting more on the colony than on individual colony members. The developmentally irreversible distinction between worker and reproductive castes makes their division of labor comparable to that of the germline and soma, and they consequently have come to be regarded as superorganisms (Wheeler, 1911; Boomsma and Gawne, 2018).
The division of non-reproductive labor among workers also becomes more specialized along the sociality gradient (Figure 1). A variety of temporal, physical, and spatial factors may direct division of non-reproductive labor (Duarte et al., 2011). Temporal subcastes, with workers transitioning from tasks within to outside the nest as they age (e.g., nurse bees transitioning to foragers), and physical subcastes with morphological distinctions exhibiting task specialization (e.g., minor and major workers and soldiers), are commonly identified across social insects (Oster and Wilson, 1978) (Figure 1). Superorganismal clades exhibit the highest degree of specialization among workers, where their cohesive actions constitute colony-level adaptations for resource acquisition, comparable to the specialized functions of somatic tissues in a multicellular organism (Wheeler, 1911; Boomsma and Gawne, 2018).
Current Knowledge on Caste-Distinct Gut Microbiomes
Microbial symbioses and division of labor are well-known to enhance resource acquisition, but few studies have compared gut microbiomes of castes and subcastes (Figure 1). In some social insect clades, the reproductive caste microbiome is distinct and drastically simplified compared to that of the worker caste. Foraging lower termite reproductives generally lack symbiotic protists that dominate the worker guts (Shimada et al., 2013; Inagaki and Matsuura, 2016), and the reproductive caste of Termitidae termites shows reduced diversity and disparate bacterial community composition (Otani et al., 2019) compared to the worker caste (Dietrich et al., 2014; Otani et al., 2014, 2016, 2019). Honey bee queens also host a simplified bacterial community that is significantly reduced compared to workers (Kapheim et al., 2015; Tarpy et al., 2015; Anderson et al., 2018). Albeit limited, data on the gut microbiomes of ants suggest some differences between reproductive and workers (Johansson et al., 2013; Brown and Wernegreen, 2016). The gut microbiomes of social wasps remain understudied, although some clades appear to retain consistent gut microbes (Stefanini et al., 2012, 2016; Gruber et al., 2019; Suenami et al., 2019).
Differences between worker subcastes have almost exclusively been explored in termites and bees. Studies suggest that the gut microbiome composition of worker bees varies with age and could be task-dependent, although patterns inferred are subtle (Guo et al., 2015; Anderson et al., 2018; Jones et al., 2018). Similarly, the fungus-farming subfamily of the Termitidae exhibits differences in gut bacterial composition related to food-processing by temporal subcastes (Hinze et al., 2002; Hongoh et al., 2006; Li et al., 2015, 2016). Termites also present differences among physical subcastes, where soldier microbiomes can be distinct from other subcastes (Hongoh et al., 2006; Shimada et al., 2013; Inagaki and Matsuura, 2016; Otani et al., 2019, but see Berlanga et al., 2011), and in highly polymorphic termites, such as the genus Macrotermes, minor and major workers and soldiers display variable microbiomes (Hongoh et al., 2006; Schnorr et al., 2019).
Sociality Supports Persistent Symbiotic Gut Microbiota
For an individual insect, the repeated remodeling of the gut and shedding of the microbiota throughout development can impede extracellular gut symbionts from persisting within individuals and over generations (Engel and Moran, 2013). Group living overcomes this challenge by providing access to microbiota through social interactions (Troyer, 1984; Nalepa et al., 2001; Lombardo, 2008; Engel and Moran, 2013).
Microbiota characteristic of natal social insect colonies have been suggested to be transmitted by reproductives in colonies founded independently (Benjamino and Graf, 2016; Meirelles et al., 2016; Stefanini et al., 2016; Diouf et al., 2018), or by workers and reproductives (Kwong et al., 2014) in colonies that are founded dependently, such as honey bees and army ants. Workers within colonies then stabilize the gut microbiota through intracolonial transfer of symbionts; newly-eclosed microbe-free workers typically receive inocula from mature workers through fecal-oral transmission (Wheeler, 1984; Ohkuma and Brune, 2010; Powell et al., 2014; Lanan et al., 2016). Additional oral exchanges, interactions, and the shared nest environment and resources facilitate continuous microbiota transmission and homogenization between colony members (Martinson et al., 2012; Stefanini et al., 2012; Powell et al., 2014; Zhukova et al., 2017). The reliable transmission of gut microbes between and within colonies thus allows transgenerational persistence of microbial communities.
Host transmission of heritable extracellular symbionts has been hypothesized to result in long-term associations between some social insects and specialized microbiota. Termites, social corbiculate bees, and clades of ants consistently host microbial phylotypes over evolutionary timescales, as indicated by patterns of phylogenetic congruence of microbial communities with hosts (Russell et al., 2009; Dietrich et al., 2014; Sanders et al., 2014; Kwong et al., 2017; Lukasik et al., 2017; Bourguignon et al., 2018; Sapountzis et al., 2019). Although this indicates vertical transmission across generations, most symbioses are characterized by the presence of at least some degree of host switching, suggesting that horizontal transmission persists across host clades (Koch et al., 2013; Sanders et al., 2014; Kwong et al., 2017; Bourguignon et al., 2018). Nevertheless, the extensive evolutionary histories of hosts and symbionts are presumably facilitated by social interactions and insect physiological or morphological characteristics that promote specific microbial partners (c.f. Kwong and Moran, 2015; Lanan et al., 2016; Sapountzis et al., 2019).
Discussion
Is the Colony Gut Microbiota Optimized Through Social Organization?
We hypothesize that hosts may organize and optimize the gut microbiota among colony members to increase the benefits of symbioses and social organization. Consistent caste-specific differences in honey bee and termite microbiomes suggest that microbial community compositions have been selected over evolutionary time to align with caste roles. If these caste-specific microbiomes enhance productivity of symbioses and efficacy of division of labor, they may be selected for as an emergent property of the combined effects of division of caste and microbiome labor.
Hosts are under strong selection to promote a beneficial microbiota; particularly in semi-closed systems like the gut (Foster et al., 2017). While microbes may compete within the gut ecosystem, potentially reducing symbiont expression of cooperative traits, hosts wrangle microbiota into stable and productive communities (Frank, 1996; Coyte et al., 2015). Hosts may shape microbiomes through mechanisms that regulate the immigration of microbes, support specific microbial community members through immune, physiological or dietary responses, and compartmentalize microbial communities (Frank, 1996; Foster et al., 2017), which in the social insects can occur both at the level of individuals and the colony. This could increase the net benefit of symbioses because optimization of microbiomes within specific castes may promote a simplified microbiota to meet distinct needs, enhancing the productivity of individual communities, while the colony-level conglomerate remains diverse.
Conclusions about optimization of symbiotic communities require elucidation of their compositions, functions, and impacts on host reproductive success. Since some microbes within a community could be driven by diet, gut morphology and physiochemistry, social interactions, or environmental exposure without further implications to the host, we propose three necessary levels of evidence to elucidate symbiotic benefit: (a) identification of consistent microbial communities within clades and castes, (b) characterization of conserved functions and benefits to hosts, and (c) determination of how hosts select for microbial community compositions and functions. With these needs in mind, we propose three focal areas that we believe can provide insight into the implications of structured colony microbiomes.
Caste Microbiomes Along the Sociality Gradient
The distinctiveness of caste microbiomes should predictably increase along the sociality gradient. In lower social grades, all colony members remain totipotent and typically undergo physical remodeling when transitioning to a different caste (Roisin, 1990; Sumner et al., 2010), which may prevent strong specialization of their gut microbiota. In contrast, early developmental determination of committed castes may fine-tune the composition of microbial communities over their lifetime to meet divergent needs, resulting in distinct microbiota structure, and function. Termites and wasps are promising models to test this prediction as they have representative clades across the sociality gradient (Figure 1).
Social structure constrains the level of adaptation and thus the potential disparity in caste and subcaste microbiota. In social grades that have not undergone the major transition to superorganismality, the distinct microbiomes may represent individual-level adaptations, and host interests in direct fitness benefits may limit specialization (Boomsma and Gawne, 2018; Cooper and West, 2018). After the major transition, however, workers can only improve their reproductive success through indirect fitness benefits gained at the colony level; thus, the colony acts as a fitness maximizing agent that can develop group-level adaptations (Gardner and Grafen, 2009), such as distinct caste and subcaste microbiota that optimize resource use and maximize colony fitness.
Colony and Microbiota Development
The microbiomes of organisms are characterized by shifts associated with changing physiology and needs over the lifetime of the individual, and social insect colonies likely undergo comparable processes, from founding (possibly excluding taxa with dependent founding) through growth and development to maturity (left part of Figure 2). If caste specificities in microbiomes play important roles for colony reproductive success, the characterization of their development over colony maturation, during which caste roles are established and maintained, may allow us to corroborate functional predictions aligned with the division of labor.
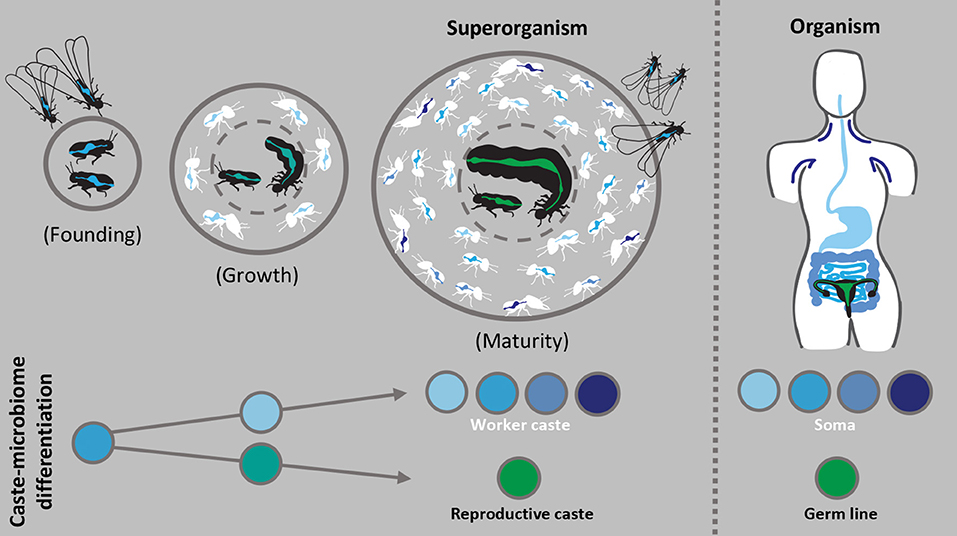
Figure 2. Caste-specific microbiomes during growth and maturity of the superorganism. The left panel illustrates how gut microbiomes (spectrum of blue to green) of the reproductive (black) and worker caste (white) may structurally and functionally differentiate during the development of an independently founded colony. Such colonies undergo ergonomic growth before reaching maturity, during which caste roles establish and gut microbiota may similarly diverge. Reproductive gut microbiota (green) likely simplifies in structure and function while worker microbiota (blue) remains relatively diverse and may be partitioned among subcastes. The right panel proposes that the general structure and function of the superorganism microbiota could be analogous to that of multicellular organisms, in this case microbiota of human tissues. The major transition to superorganismality causes selection to act on the colony as a unit, similar to an individual organism, and the binary distinction between the worker and reproductive caste is comparable to the soma and germ line. Greater understanding of the cohesive actions carried out by the divisible units of superorganisms and their microbial partners may allow us to derive novel insights into higher-level emergent characteristics typically associated with and observed in organisms.
As colonies grow, workers liberate reproductives from non-reproductive tasks, and both worker and reproductive castes become increasingly specialized (Wheeler, 1911; Oster and Wilson, 1978). Similarly, we predict that the structure and function of worker and reproductive gut microbiomes diverge during colony growth (Figure 2). The gut microbiomes of termite reproductives indicate a transition from diverse to simplified communities from founding to maturity (Shimada et al., 2013; Benjamino and Graf, 2016; Inagaki and Matsuura, 2016; Diouf et al., 2018; Otani et al., 2019) (Figure 2), which has been suggested to be a consequence of specialization on reproduction and dependence on workers (Chouvenc and Su, 2017). Similarly, the gut microbiome of honey bee queens exhibits slight changes with age (Tarpy et al., 2015; Anderson et al., 2018), suggesting that physiological and dietary changes related to reproductive specialization and duration may influence gradual shifts the reproductive microbiota. Additionally, as the number of workers increases, temporal and physical subcastes may develop unique microbiota, where age-dependent tasks, environments, and diets could influence microbial community composition. Exploration of changes in host characteristics and microbiota during colony development could thus clarify mechanisms of selection and caste complementarity, while examining changes in the functional microbiome and its influence on colony growth may elucidate how a divided microbiome supports colony maturation and the accretion of fitness benefits. Secondarily, the significance of independent or dependent colony founding should explored, as it independent founding may cause a microbial bottleneck that reduces both the stability and diversity of the colony gut microbiome (Kwong et al., 2017; Lukasik et al., 2017). Consistent with our previous inferences, we expect that insights into these processes can be achieved through comparative analyses of social insects across the spectrum of social organization.
The Structured Gut Microbiota of the Superorganism
We contend that the tightest associations and most distinct caste-specific microbiomes will be in clades with extreme and irreversible division of labor, the superorganisms. In particular, social insects such as honey bees, certain ants and the Termitidae may provide the greatest insights because the complex division of non-reproductive labor supports remarkable fecundity of reproductives (Winston, 1991; Kaib et al., 2001; Hölldobler and Wilson, 2009). A simple reproductive gut microbiota may be influenced by traits of the individual, such as their unique diet, typically consisting of buccal secretions from workers (Noirot, 1969; Haydak, 1979), and limited exposure to environmental microbes (Cremer et al., 2018; Stroeymeyt et al., 2018), thus, reducing microbial immigration. The reproductive microbiome then likely supports specific metabolic demands, critical disease defense, or increased longevity, while most other colony functions are outsourced to workers and their microbiomes (Figure 2).
Complexity in the worker gut microbiota is likely driven by variable host tasks, diets, and interactions. Immigration of microbiota into the worker gut is controlled during post-eclosion microbial inoculation (Ohkuma and Brune, 2010; Powell et al., 2014; Lanan et al., 2016) and remodeling of gut morphology (Zhukova et al., 2017). Changes in host diet and physiology, such as the differential diet and gut enzyme expression of honey bee workers with age (Crailsheim et al., 1992), may influence the gut microbiota. Subcaste microbiota could additionally be influenced by interaction and transmission networks, resulting in compartmentalization. Notably, since the majority of microbiota studies utilize 16S rRNA profiling, it is likely that compositional differences at the bacterial strain level have been overlooked (Engel et al., 2014; Ellegaard and Engel, 2016), as indicated by compartmentalization of strains between individual honey bees and variable functional gene content across age groups (Ellegaard and Engel, 2019). Compartmentalization among subcastes would conceivably allow microbes to cater to specific host roles, create adaptive heterogeneity (Masuda et al., 2015; Kennedy et al., 2017), and reduce the potential for competition between symbionts (Frank, 1996). Consequently, the various roles and interactions of worker (sub)caste(s) should promote functionally diverse microbiomes that hosts optimally divide to complement tasks.
A superorganism's highly structured colony microbiome, shaped by distinct (sub)caste communities, may be adaptive to the colony as a unit, similar to the composite of different microbiomes observed in tissues or body regions of multicellular organisms such as humans (The Human Microbiome Consotrium, 2012) (Figure 2). The major evolutionary transition to superorganismal social complexity shifts selection to act more strongly at the colony-level, similar to selection on multicellular organisms, and the coherent actions of morphologically committed reproductive and non-reproductive (sub)castes is comparable to the division of cells into germ and soma (Wheeler, 1911; Boomsma and Gawne, 2018). The analogous organization of the two systems allows comparisons and insights into higher-level characteristics typically associated with and observed in organisms (Helanterä, 2016; Kennedy et al., 2017), which could include the structure and function of the microbiome (Figure 2). For example, the significance of microbial partners in the framework of social immunity, another emergent property enabled by social structure (Cremer et al., 2007, 2018), may be compared to the interplay between microbiota and the human immune system (Hooper et al., 2012). We believe that considering this analogy in future research will help improve our fundamental understanding of the impact of symbioses on individuals, (sub)castes and superorganismal division of labor.
Conclusion
Current research on social insect symbioses focuses on microbiome function and the importance of social life in maintaining a consistent microbiota. However, we have yet to fully integrate these paradigms. The division of labor, which directs social roles and interactions, may secondarily partition the microbiota, shaping caste-specific microbiomes that dually enhance productivity of symbioses and efficiency of castes. While individual and colony-level mechanisms may drive distinctions in gut microbiota, microbial interactions within these communities should be examined to fully understand structured microbiota as a potential adaptation. Overall, we contend that integration of gut symbionts into the framework of sociality defined by division of labor may elucidate its potential adaptive value to individual insects and the colony as a whole.
Author Contributions
VS, JR-H, and MP conceived the ideas. VS drafted the first version of text and figures. VS, JR-H, BT, KE, PS, MV-C, and MP contributed with expertise, input, and edits throughout the remainder of the text.
Funding
This work was supported by a Ph.D. stipend from the Department of Biology, University of Copenhagen to VS, a Ph.D. studentship from the Natural Environment Research Council (NERC) to BT, HFSP-RGP0060/2018 to MV-C, and an ERC Consolidator grant (771349) to MP.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Jacobus J. Boomsma and E. Allen Herre for fruitful discussion and Kasun H. Bodawatta for comments to an earlier version of the manuscript.
References
Aizen, M. A., Garibaldi, L. A., Cunningham, S. A., and Klein, A. M. (2008). Long-term global trends in crop yield and production reveal no current pollination shortage but increasing pollinator dependency. Curr. Biol. 18, 1572–1575. doi: 10.1016/j.cub.2008.08.066
Anderson, C., and Ratnieks, F. L. W. (1999). Task partitioning in insect societies. I. Effect of colony size on queueing delay and colony ergonomic efficiency. Am. Nat. 154, 521–535. doi: 10.1086/303255
Anderson, K. E., Ricigliano, V. A., Mott, B. M., Copeland, D. C., Floyd, A. S., and Maes, P. (2018). The queen's gut refines with age: longevity phenotypes in a social insect model. Microbiome 6:108. doi: 10.1186/s40168-018-0489-1
Benjamino, J., and Graf, J. (2016). Characterization of the core and caste-specific microbiota in the termite, Reticulitermes flavipes. Front. Microbiol. 7:171. doi: 10.3389/fmicb.2016.00171
Berlanga, M., Paster, B. J., Grandcolas, P., and Guerrero, R. (2011). Comparison of the gut microbiota from soldier and worker castes of the termite Reticulitermes grassei. Int. Microbiol. 14, 83–93. doi: 10.2436/20.1501.01.138
Bignell, D. E., and Eggleton, P. (2000). “Termites in ecosystems,” in Termites: Evolution, Sociality, Symbioses, Ecology, eds T. Abe, D. E. Bignell, and M. Higashi (Dordecht: Springer), 363–387. doi: 10.1007/978-94-017-3223-9_17
Boomsma, J. J., and Gawne, R. (2018). Superorganismality and caste differentiation as points of no return: how the major evolutionary transitions were lost in translation. Biol. Rev. Camb. Philos. Soc. 93, 28–54. doi: 10.1111/brv.12330
Bourguignon, T., Lo, N., Dietrich, C., Sobotnik, J., Sidek, S., Roisin, Y., et al. (2018). Rampant host switching shaped the termite gut microbiome. Curr. Biol. 28, 649–654.e642. doi: 10.1016/j.cub.2018.01.035
Brown, B. P., and Wernegreen, J. J. (2016). Deep divergence and rapid evolutionary rates in gut-associated Acetobacteraceae of ants. BMC Microbiol. 16:140. doi: 10.1186/s12866-016-0721-8
Brune, A. (2014). Symbiotic digestion of lignocellulose in termite guts. Nat. Rev. Microbiol. 12, 168–180. doi: 10.1038/nrmicro3182
Chouvenc, T., and Su, N. Y. (2017). Irreversible transfer of brood care duties and insights into the burden of caregiving in incipient subterranean termite colonies. Ecol. Entomol. 42, 777–784. doi: 10.1111/een.12443
Cleveland, L. R. (1923). Symbiosis between termites and their intestinal protozoa. Proc. Natl. Acad. Sci. U.S.A. 9, 424–428. doi: 10.1073/pnas.9.12.424
Cooper, G. A., and West, S. A. (2018). Division of labour and the evolution of extreme specialization. Nat. Ecol. Evol. 2, 1161–1167. doi: 10.1038/s41559-018-0564-9
Coyte, K. Z., Schluter, J., and Foster, K. R. (2015). The ecology of the microbiome: networks, competition, and stability. Science 350, 663–666. doi: 10.1126/science.aad2602
Crailsheim, K., Schneider, L. H. W., Hrassnigg, N., Bühlmann, G., Brosch, U., Gmeinbauer, R., et al. (1992). Pollen consumption and utilization in worker honey bees (Apis mellifera carnica): dependence on individual age and function. J. Insect. Physiol. 38, 409–419. doi: 10.1016/0022-1910(92)90117-V
Cremer, S., Armitage, S. A., and Schmid-Hempel, P. (2007). Social immunity. Curr. Biol. 17, R693–702. doi: 10.1016/j.cub.2007.06.008
Cremer, S., Pull, C. D., and Furst, M. A. (2018). Social immunity: emergence and evolution of colony-level disease protection. Annu. Rev. Entomol. 63, 105–123. doi: 10.1146/annurev-ento-020117-043110
Del Toro, I., Ribbons, R. R., and Pelini, S. L. (2012). The little things that run the world revisited: a review of ant-mediated ecosystem services and disservices (Hymenoptera: Formicidae). Myrmecol. News 17, 133–146.
Dietrich, C., Kohler, T., and Brune, A. (2014). The cockroach origin of the termite gut microbiota: patterns in bacterial community structure reflect major evolutionary events. Appl. Environ. Microbiol. 80, 2261–2269. doi: 10.1128/AEM.04206-13
Diouf, M., Herve, V., Mora, P., Robert, A., Frechault, S., Rouland-Lefevre, C., et al. (2018). Evidence from the gut microbiota of swarming alates of a vertical transmission of the bacterial symbionts in nasutitermes arborum (Termitidae, Nasutitermitinae). Antonie van Leeuwenhoek 111, 573–587. doi: 10.1007/s10482-017-0978-4
Du, H., Chouvenc, T., and Su, N. Y. (2017). Development of age polyethism with colony maturity in coptotermes formosanus (Isoptera: Rhinotermitidae). Environ. Entomol. 46, 311–318. doi: 10.1093/ee/nvw162
Duarte, A., Weissing, F. J., Pen, I., and Keller, L. (2011). An evolutionary perspective on self-organized division of labor in social insects. Annu. Rev. Ecol. Evol. Syst. 42, 91–110. doi: 10.1146/annurev-ecolsys-102710-145017
Ellegaard, K. M., and Engel, P. (2016). Beyond 16S rRNA community profiling: intra-species diversity in the gut microbiota. Front. Microbiol. 7:1475. doi: 10.3389/fmicb.2016.01475
Ellegaard, K. M., and Engel, P. (2019). Genomic diversity landscape of the honey bee gut microbiota. Nat. Commun. 10:446. doi: 10.1038/s41467-019-08303-0
Engel, P., and Moran, N. A. (2013). The gut microbiota of insects - diversity in structure and function. FEMS Microbiol. Rev. 37, 699–735. doi: 10.1111/1574-6976.12025
Engel, P., Stepanauskas, R., and Moran, N. A. (2014). Hidden diversity in honey bee gut symbionts detected by single-cell genomics. PLoS Genet. 10:e1004596. doi: 10.1371/journal.pgen.1004596
Evans, T. A. (2006). Foraging and building in subterranean termites: task switchers or reserve labourers. Insectes Soc. 53, 56–64. doi: 10.1007/s00040-005-0835-8
Foster, K. R., Schluter, J., Coyte, K. Z., and Rakoff-Nahoum, S. (2017). The evolution of the host microbiome as an ecosystem on a leash. Nature 548, 43–51. doi: 10.1038/nature23292
Frank, S. A. (1996). Host-symbiont conflict over the mixing of symbiotic lineages. Proc. Biol. Sci. 263, 339–344. doi: 10.1098/rspb.1996.0052
Gardner, A., and Grafen, A. (2009). Capturing the superorganism: a formal theory of group adaptation. J. Evol. Biol. 22, 659–671. doi: 10.1111/j.1420-9101.2008.01681.x
Goulson, D., Peat, J., Stout, J. C., Tucker, J., Darvill, B., Derwent, L. C., et al. (2002). Can alloethism in workers of the bumblebee, Bombus terrestris, be explained in terms of foraging efficiency? Anim. Behav. 64, 123–130. doi: 10.1006/anbe.2002.3041
Gruber, M. A., Quinn, O., Baty, J. W., Dobelmann, J., Haywood, J., et al. (2019). Fitness and microbial networks of the common wasp, Vespula vulgaris (Hymenoptera: Vespidae), in its native and introduced ranges. Ecol. Entomol. 44, 512–523. doi: 10.1111/een.12732
Grüter, C., Segers, F. H., Menezes, C., Vollet-Neto, A., Falcon, T., von Zuben, L., et al. (2017). Repeated evolution of soldier sub-castes suggests parasitism drives social complexity in stingless bees. Nat. Commun. 8:4. doi: 10.1038/s41467-016-0012-y
Guo, J., Wu, J., Chen, Y., Evans, J. D., Dai, R., Luo, W., et al. (2015). Characterization of gut bacteria at different developmental stages of Asian honey bees, Apis cerana. J. Invertebr. Pathol. 127, 110–114. doi: 10.1016/j.jip.2015.03.010
Haydak, M. H. (1979). Honey bee nutrition. Annu. Rev. Entomol. 15, 143–156. doi: 10.1146/annurev.en.15.010170.001043
Helanterä, H. (2016). An organismal perspective on the evolution of insect societies. Front. Ecol. Evol. 4:1–12. doi: 10.3389/fevo.2016.00006
Hinze, B., Crailsheim, K., and Leuthold, R. H. (2002). Polyethism in food processing and social organisaiton in the nest of Macrotermes bellicosus (Isoptera, Termitidae). Insectes Soc. 49, 31–37. doi: 10.1007/s00040-002-8275-1
Hölldobler, B., and Wilson, E. O. (2009). The Superorgansim: The Beauty, Elegance, and Strangeness of Insect Societies. New York, NY: W. W Norton & Company.
Hongoh, Y., Ekpornprasit, L., Inoue, T., Moriya, S., Trakulnaleamsai, S., Ohkuma, M., et al. (2006). Intracolony variation of bacterial gut microbiota among castes and ages in the fungus-growing termite Macrotermes gilvus. Mol. Ecol. 15, 505–516. doi: 10.1111/j.1365-294X.2005.02795.x
Hooper, L. V., Littman, D. R., and Macpherson, A. J. (2012). Interactions between the microbiota and the immune system. Science 336, 1268–1273. doi: 10.1126/science.1223490
Hu, Y., Sanders, J. G., Lukasik, P., D'Amelio, C. L., Millar, J. S., Vann, D. R., et al. (2018). Herbivorous turtle ants obtain essential nutrients from a conserved nitrogen-recycling gut microbiome. Nat. Commun. 9:964. doi: 10.1038/s41467-018-04935-w
Inagaki, T., and Matsuura, K. (2016). Colony-dependent sex differences in protozoan communities of the lower termite Reticulitermes speratus (Isoptera: Rhinotermitidae). Ecol. Res. 31, 749–755. doi: 10.1007/s11284-016-1387-2
Inagaki, T., and Matsuura, K. (2018). Extended mutualism between termites and gut microbes: nutritional symbionts contribute to nest hygiene. Naturwissenschaften 105:52. doi: 10.1007/s00114-018-1580-y
Jandt, J. M., and Toth, A. L. (2015). “Physiological and genomic mechanisms of social organization in wasps (Family: Vespidae),” in Advances in Insect Physiology, eds A. Zayed and C. F. Kent (London: Academic Press), 95–130. doi: 10.1016/bs.aiip.2015.01.003
Johansson, H., Dhaygude, K., Lindstrom, S., Helantera, H., Sundstrom, L., and Trontti, K. (2013). A metatranscriptomic approach to the identification of microbiota associated with the ant Formica exsecta. PLoS ONE 8:e79777. doi: 10.1371/journal.pone.0079777
Jones, J. C., Fruciano, C., Marchant, J., Hildebrand, F., Forslund, S., Bork, P., et al. (2018). The gut microbiome is associated with behavioural task in honey bees. Insectes Soc. 65, 419–429. doi: 10.1007/s00040-018-0624-9
Kaib, M., Hacker, M., and Brandl, R. (2001). Egg-laying in monogynous and polygynous colonies of the termite Macrotermes michaelseni (Isoptera, Macrotermitidae). Insectes Soc. 48, 231–237. doi: 10.1007/PL00001771
Kapheim, K. M., Rao, V. D., Yeoman, C. J., Wilson, B. A., White, B. A., Goldenfeld, N., et al. (2015). Caste-specific differences in hindgut microbial communities of honey bees (Apis mellifera). PLoS ONE 10:e0123911. doi: 10.1371/journal.pone.0123911
Kennedy, P., Baron, G., Qiu, B., Freitak, D., Helanterä, H., Hunt, E. R., et al. (2017). Deconstructing superorganisms and societies to address big questions in biology. Trends Ecol. Evol. 32, 861–872. doi: 10.1016/j.tree.2017.08.004
Kesnerova, L., Mars, R. A. T., Ellegaard, K. M., Troilo, M., Sauer, U., and Engel, P. (2017). Disentangling metabolic functions of bacteria in the honey bee gut. PLoS Biol. 15:e2003467. doi: 10.1371/journal.pbio.2003467
Koch, H., Abrol, D. P., Li, J., and Schmid-Hempel, P. (2013). Diversity and evolutionary patterns of bacterial gut associates of corbiculate bees. Mol. Ecol. 22, 2028–2044. doi: 10.1111/mec.12209
Koch, H., and Schmid-Hempel, P. (2011). Socially transmitted gut microbiota protect bumble bees against an intestinal parasite. Proc. Natl. Acad. Sci. U.S.A. 108, 19288–19292. doi: 10.1073/pnas.1110474108
Korb, J., and Thorne, B. (2017). “Sociality in Termites,” in Comparative Social Evolution, eds. D. R. Rubenstein, and P. Abbot (Cambridge, UK: Cambridge University Press).
Kwong, W. K., Engel, P., Koch, H., and Moran, N. A. (2014). Genomics and host specialization of honey bee and bumble bee gut symbionts. Proc. Natl. Acad. Sci. U.S.A. 111, 11509–11514. doi: 10.1073/pnas.1405838111
Kwong, W. K., Medina, L. A., Koch, H., Sing, K. W., Soh, E. J. Y., Ascher, J. S., et al. (2017). Dynamic microbiome evolution in social bees. Sci. Adv. 3:e1600513. doi: 10.1126/sciadv.1600513
Kwong, W. K., and Moran, N. A. (2015). Evolution of host specialization in gut microbes: the bee gut as a model. Gut Microbes 6, 214–220. doi: 10.1080/19490976.2015.1047129
Kwong, W. K., and Moran, N. A. (2016). Gut microbial communities of social bees. Nat. Rev. Microbiol. 14, 374–384. doi: 10.1038/nrmicro.2016.43
Lanan, M. C., Rodrigues, P. A., Agellon, A., Jansma, P., and Wheeler, D. E. (2016). A bacterial filter protects and structures the gut microbiome of an insect. ISME J. 10, 1866–1876. doi: 10.1038/ismej.2015.264
Li, H., Dietrich, C., Zhu, N., Mikaelyan, A., Ma, B., Pi, R., et al. (2016). Age polyethism drives community structure of the bacterial gut microbiota in the fungus-cultivating termite Odontotermes formosanus. Environ. Microbiol. 18, 1440–1451. doi: 10.1111/1462-2920.13046
Li, H., Yang, M., Chen, Y., Zhu, N., Lee, C. Y., Wei, J. Q., et al. (2015). Investigation of age polyethism in food processing of the fungus-growing termite Odontotermes formosanus (Blattodea: Termitidae) using a laboratory artificial rearing system. J. Econ. Entomol. 108, 266–273. doi: 10.1093/jee/tou005
Lombardo, M. P. (2008). Access to mutualistic endosymbiotic microbes: an underappreciated benefit of group living. Behav. Ecol. Sociobiol. 62, 479–498. doi: 10.1007/s00265-007-0428-9
Lukasik, P., Newton, J. A., Sanders, J. G., Hu, Y., Moreau, C. S., Kronauer, D. J. C., et al. (2017). The structured diversity of specialized gut symbionts of the New World army ants. Mol. Ecol. 26, 3808–3825. doi: 10.1111/mec.14140
Martinson, V. G., Moy, J., and Moran, N. A. (2012). Establishment of characteristic gut bacteria during development of the honeybee worker. Appl. Environ. Microbiol. 78, 2830–2840. doi: 10.1128/AEM.07810-11
Masuda, N., O'Shea-Wheller, T. A., Doran, C., and Franks, N. R. (2015). Computational model of collective nest selection by ants with heterogeneous acceptance thresholds. R. Soc. Open. Sci. 2:140533. doi: 10.1098/rsos.140533
Maynard Smith, J., and Szathmáry, E. (1995). The Major Transitions in Evolution. New York, NY: Oxford University Press.
Meirelles, L. A., McFrederick, Q. S., Rodrigues, A., Mantovani, J. D., de Melo Rodovalho, C., Ferreira, H., et al. (2016). Bacterial microbiomes from vertically transmitted fungal inocula of the leaf-cutting ant Atta texana. Environ. Microbiol. Rep. 8, 630–640. doi: 10.1111/1758-2229.12415
Nalepa, C. A., Bignell, D. E., and Bandi, C. (2001). Detrivory, coprophagy, and the evolution of digestive mutualisms in Dictyoptera. Insectes Soc. 48, 194–201. doi: 10.1007/PL00001767
Noirot, C. (1969). “Glands and secretions,” in Biology of Termites, eds K. Krishna, and F. Weesner (New York, NY: Academic Press), 89–123.
Ohkuma, M., and Brune, A. (2010). “Diversity, structure, and evolution of the termite gut microbial community,” in Biology of Termites: a Modern Synthesis, eds D. E. Bignell, Y. Roisin, and N. Lo (Dorderecht: Springer), 413–438. doi: 10.1007/978-90-481-3977-4_15
Oster, G. F., and Wilson, E. O. (1978). Caste and Ecology in the Social Insects. (Princeton, NJ: Princeton University Press).
Otani, S., Hansen, L. H., Sorensen, S. J., and Poulsen, M. (2016). Bacterial communities in termite fungus combs are comprised of consistent gut deposits and contributions from the environment. Microb. Ecol. 71, 207–220. doi: 10.1007/s00248-015-0692-6
Otani, S., Mikaelyan, A., Nobre, T., Hansen, L. H., Kone, N. A., Sorensen, S. J., et al. (2014). Identifying the core microbial community in the gut of fungus-growing termites. Mol. Ecol. 23, 4631–4644. doi: 10.1111/mec.12874
Otani, S., Zhukova, M., Kone, N. A., da Costa, R. R., Mikaelyan, A., Sapountzis, P., et al. (2019). Gut microbial compositions mirror caste-specific diets in a major lineage of social insects. Environ. Microbiol. Rep. 11, 196–205. doi: 10.1111/1758-2229.12728
Peterson, B. F., and Scharf, M. E. (2016). Lower termite associations with microbes: synergy, protection, and interplay. Front. Microbiol. 7:422. doi: 10.3389/fmicb.2016.00422
Powell, J. E., Martinson, V. G., Urban-Mead, K., and Moran, N. A. (2014). Routes of acquisition of the gut microbiota of the honey bee Apis mellifera. Appl. Environ. Microbiol. 80, 7378–7387. doi: 10.1128/AEM.01861-14
Raymann, K., Shaffer, Z., and Moran, N. A. (2017). Antibiotic exposure perturbs the gut microbiota and elevates mortality in honeybees. PLoS Biol. 15:e2001861. doi: 10.1371/journal.pbio.2001861
Richter, M. R. (2000). Social wasp (Hymenoptera: Vespidae) foraging behavior. Annu. Rev. Entomol. 45, 121–150. doi: 10.1146/annurev.ento.45.1.121
Roisin, Y. (1990). Queen replacement in the termite Microcerotermes papuanus. Entomol. Exp. Appl. 56, 83–90. doi: 10.1111/j.1570-7458.1990.tb01383.x
Russell, J. A., Moreau, C. S., Goldman-Huertas, B., Fujiwara, M., Lohman, D. J., and Pierce, N. E. (2009). Bacterial gut symbionts are tightly linked with the evolution of herbivory in ants. Proc. Natl. Acad. Sci. U.S.A. 106, 21236–21241. doi: 10.1073/pnas.0907926106
Sanders, J. G., Powell, S., Kronauer, D. J., Vasconcelos, H. L., Frederickson, M. E., and Pierce, N. E. (2014). Stability and phylogenetic correlation in gut microbiota: lessons from ants and apes. Mol. Ecol. 23, 1268–1283. doi: 10.1111/mec.12611
Sapountzis, P., Nash, D. R., Schiott, M., and Boomsma, J. J. (2019). The evolution of abdominal microbiomes in fungus-growing ants. Mol. Ecol. 28, 879–899. doi: 10.1111/mec.14931
Sapountzis, P., Zhukova, M., Shik, J. Z., Schiott, M., and Boomsma, J. J. (2018). Reconstructing the functions of endosymbiotic Mollicutes in fungus-growing ants. Elife 7:e39209. doi: 10.7554/eLife.39209.037
Schnorr, S. L., Hofman, C. A., Netshifhefhe, S. R., Duncan, F. D., Honap, T. P., Lesnik, J., et al. (2019). Taxonomic features and comparisons of the gut microbiome from two edible fungus-farming termites (Macrotermes falciger; M. natalensis) harvested in the Vhembe district of Limpopo, South Africa. BMC Microbiol. 19:164. doi: 10.1186/s12866-019-1540-5
Seeley, T. D. (1982). Adaptive significance of the age polyethism schedule in honey bee colonies. Behav. Ecol. Sociobiol. 11, 41287–293. doi: 10.1007/BF00299306
Shimada, K., Lo, N., Kitade, O., Wakui, A., and Maekawa, K. (2013). Cellulolytic protist numbers rise and fall dramatically in termite queens and kings during colony foundation. Eukaryotic Cell 12, 545–550. doi: 10.1128/EC.00286-12
Spaethe, J., and Weidenmüller, A. (2002). Size variation and foraging rate in bumblebees (Bombus terrestris). Insect. Soc. 49, 142–146. doi: 10.1007/s00040-002-8293-z
Stefanini, I., Dapporto, L., Berna, L., Polsinelli, M., Turillazzi, S., and Cavalieri, D. (2016). Social wasps are a Saccharomyces mating nest. Proc. Natl. Acad. Sci. U.S.A. 113, 2247–2251. doi: 10.1073/pnas.1516453113
Stefanini, I., Dapporto, L., Legras, J. L., Calabretta, A., Di Paola, M., De Filippo, C., et al. (2012). Role of social wasps in Saccharomyces cerevisiae ecology and evolution. Proc. Natl. Acad. Sci. U.S.A. 109, 13398–13403. doi: 10.1073/pnas.1208362109
Stroeymeyt, N., Grasse, A. V., Crespi, A., Mersch, D. P., Cremer, S., and Keller, L. (2018). Social network plasticity decreases disease transmission in a eusocial insect. Science 362, 941–945. doi: 10.1126/science.aat4793
Suenami, S., Konishi Nobu, M., and Miyazaki, R. (2019). Community analysis of gut microbiota in hornets, the largest eusocial wasps, Vespa mandarinia and V. simillima. Sci. Rep. 9:9830. doi: 10.1038/s41598-019-46388-1
Sumner, S., Kelstrup, H., and Fanelli, D. (2010). Reproductive constraints, direct fitness and indirect fitness benefits explain helping behaviour in the primitively eusocial wasp, Polistes canadensis. Proc. Biol. Sci. 277, 1721–1728. doi: 10.1098/rspb.2009.2289
Tarpy, D. R., Mattila, H. R., and Newton, I. L. (2015). Development of the honey bee gut microbiome throughout the queen-rearing process. Appl. Environ. Microbiol. 81, 3182–3191. doi: 10.1128/AEM.00307-15
Taylor, B. A., Reuter, M., and Sumner, S. (2019). Patterns of reproductive differentiation and reproductive plasticity in the major evolutionary transition to superorganismality. Curr. Opin. Insect. Sci. 34, 40–47. doi: 10.1016/j.cois.2019.02.007
The Human Microbiome Consotrium (2012). Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214. doi: 10.1038/nature11234
Troyer, K. (1984). Microbes, herbivory and the evolution of social behavior. J. Theor. Biol. 106, 157–169. doi: 10.1016/0022-5193(84)90016-X
Watson, J. A., and Sewell, J. J. (1985). “Caste development in Mastotermes and Kalotermes: which is primitive?,” in Caste Differnetiation in Social Insects, eds J.A. Watson, B. M. Okot-kotber, and C. H. Noirot (Oxford: Pergamon), 27–40. doi: 10.1016/B978-0-08-030783-1.50008-2
West, S. A., Fisher, R. M., Gardner, A., and Kiers, E. T. (2015). Major evolutionary transitions in individuality. Proc. Natl. Acad. Sci. U.S.A. 112, 10112–10119. doi: 10.1073/pnas.1421402112
Wheeler, D. E. (1984). Behavior of the ant, Procryptocerus scabriusculus (Hymenoptera: Formicidae), with comparisons to other Cephalotines. Psyche: J. Entomol. 91, 171–192. doi: 10.1155/1984/65
Wheeler, W. M. (1911). The ant-colony as an organism. J. Morphol. 22, 307–325. doi: 10.1002/jmor.1050220206
Wille, A. (1983). Biology of the stingless bess. Annu. Rev. Entomol. 28, 41–64. doi: 10.1146/annurev.en.28.010183.000353
Zheng, H., Powell, J. E., Steele, M. I., Dietrich, C., and Moran, N. A. (2017). Honeybee gut microbiota promotes host weight gain via bacterial metabolism and hormonal signaling. Proc. Natl. Acad. Sci. U.S.A. 114, 4775–4780. doi: 10.1073/pnas.1701819114
Keywords: symbiosis, division of labor, gut microbiome, major evolutionary transition, superorganism
Citation: Sinotte VM, Renelies-Hamilton J, Taylor BA, Ellegaard KM, Sapountzis P, Vasseur-Cognet M and Poulsen M (2020) Synergies Between Division of Labor and Gut Microbiomes of Social Insects. Front. Ecol. Evol. 7:503. doi: 10.3389/fevo.2019.00503
Received: 04 September 2019; Accepted: 10 December 2019;
Published: 09 January 2020.
Edited by:
Peter H. W. Biedermann, Julius Maximilian University of Würzburg, GermanyReviewed by:
Vienna Kowallik, Okinawa Institute of Science and Technology Graduate University, JapanOgao Onchuru Thomas, Kenyatta University, Kenya
Copyright © 2020 Sinotte, Renelies-Hamilton, Taylor, Ellegaard, Sapountzis, Vasseur-Cognet and Poulsen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Veronica M. Sinotte, veronica.sinotte@bio.ku.dk
 Veronica M. Sinotte
Veronica M. Sinotte Justinn Renelies-Hamilton
Justinn Renelies-Hamilton Benjamin A. Taylor2
Benjamin A. Taylor2  Kirsten M. Ellegaard
Kirsten M. Ellegaard Panagiotis Sapountzis
Panagiotis Sapountzis Michael Poulsen
Michael Poulsen