- Departamento de Microbiología y Genética, Universidad de Salamanca, Salamanca, Spain
For a long time, it was believed that a healthy plant did not harbor any microorganisms within its tissues, as these were often considered detrimental for the plant. In the last three decades, the numbers of studies on plant microbe-interactions has led to a change in our view and we now know that many of these invisible partners are essential for the overall welfare of the plant. The application of Next Generation Sequencing techniques is a powerful tool that has permitted the detection and identification of microbial communities in healthy plants. Among the new plant microbe interactions recently reported several actinobacteria such as Micromonospora are included. Micromonospora is a Gram-positive bacterium with a wide geographical distribution; it can be found in the soil, mangrove sediments, and freshwater and marine ecosistems. In the last years our group has focused on the isolation of Micromonospora strains from nitrogen fixing nodules of both leguminous and actinorhizal plants and reported for the first time its wide distribution in nitrogen fixing nodules of both types of plants. These studies have shown how this microoganism had been largely overlooked in this niche due to its slow growth. Surprisingly, the genetic diversity of Micromonospora strains isolated from nodules is very high and several new species have been described. The current data indicate that Micromonospora saelicesensis is the most frequently isolated species from the nodular tissues of both leguminous and actinorhizal plants. Further studies have also been carried out to confirm the presence of Micromonospora inside the nodule tissues, mainly by specific in situ hybridization. The information derived from the genome of the model strain, Micromonospora lupini, Lupac 08, has provided useful information as to how this bacterium may relate with its host plant. Several strategies potentially necessary for Micromonospora to thrive in the soil, a highly competitive, and rough environment, and as an endophytic bacterium with the capacity to colonize the internal plant tissues which are protected from the invasion of other soil microbes were identified. The genome data also revealed the potential of M. lupini Lupac 08 as a plant growth promoting bacterium. Several loci involved in plant growth promotion features such as the production of siderophores, phytohormones, and the degradation of chitin (biocontrol) were also located on the genome and the functionality of these genes was confirmed in the laboratory. In addition, when several host plants species were inoculated with Micromonospora strains, the plant growth enhancing effect was evident under greenhouse conditions. Unexpectedly, a high number of plant-cell wall degrading enzymes were also detected, a trait usually found only in pathogenic bacteria. Thus, Micromonospora can be added to the list of new plant-microbe interactions. The current data indicate that this microorganism may have an important application in agriculture and other biotechnological processes. The available information is promising but limited, much research is still needed to determine which is the ecological function of Micromonospora in interaction with nitrogen fixing plants.
Introduction
Bacteria, archaea, and viruses are present in every niche present in our planet and have a great impact on other forms of life. Since the appearance of plants on Earth, their capacity to adapt to different ecosystems and their evolutionary process have inherently been associated to microorganisms (Reid and Greene, 2012).
Microbial communities present in soil account for the richest reservoir of biological diversity in our planet (Berendsen et al., 2012). Microorganisms that live in the rhizosphere, the soil region influenced by plant roots, are of great importance as this is where most plant-microbe interactions occur (Schenk et al., 2012) and this complex plant-associated microbial community is for the most part beneficial to the plant (Berendsen et al., 2012). Despite the importance of microorganisms for plants, these extremely complex microbial communities have remained largely uncharacterized mainly due to our lack of culturing most microorganisms under laboratory conditions (Schenk et al., 2012). Fortunately, our awareness of mutually beneficial relationships and their potential application in biotechnological processes is expanding, in part due to the new sequencing technologies and information derived from their use.
Microbes that interact with plants are termed rhizospheric or endophytic depending on their localization outside or inside the plant, respectively, and many endophytes originate from the rhizosphere or phyllosphere (Dudeja et al., 2012). These organisms can accelerate seed germination, promote plant establishment under adverse conditions, enhance plant growth or prevent pathogen infections (Hurek et al., 2002; Ryan et al., 2008). Thus, a complex and invisible ecosystem sustains plant growth and health (Reid and Greene, 2012). The potential application of beneficial microbes in different fields (e.g., agriculture, biotechnology, medicine, etc.) is immense provided progress is made in understanding these complex plant-microbe interactions in a global context.
Hitherto, plant associated Gram-negative bacteria are the best studied given their relative facility to be recovered from internal plant tissues and also because mutants can be easily generated for interaction studies (Francis et al., 2010). However, many Gram-positive bacteria included in the phyla Firmicutes and Actinobacteria (e.g., Bacillus, Micromonospora, Streptomyces, etc.) have excellent biocontrol, plant growth-promoting and bioremediation activities. In addition, several characteristics observed including pigment and spore production, biosynthesis of secondary metabolites and unique lifestyles present in these microorganisms can be advantageous for different biotechnological applications, including agriculture.
In this review, the diversity and interaction between actinobacteria and plants will be discussed, focusing on their ecological aspects and potential applications in agriculture. The second part of this revision will focus on the specific interaction of the genus Micromonospora with nitrogen fixing plants.
Plant-Associated Actinobacteria
Actinobacteria represent approximately 20–30% of the rhizospheric microbial community (Bouizgarne and Ben Aouamar, 2014). They are Gram-positive and show a wide morphological spectrum ranging from unicellular organisms to branching filaments that form a mycelium. A unique feature is their high guanine plus cytosine content (>50%) in their genome. These microorganisms are for the most part saprophytic, soil-dwelling organisms with an important role in the turnover of organic matter. In addition, many species are sporulated and spend the majority of their life cycles as semidormant spores (Coombs and Franco, 2003a).
Several taxa are well-known to interact with plants and these include examples of both endophytic and plant-pathogenic species. The first actinobacterial endophyte isolated, Frankia (Callaham et al., 1978), is a nitrogen-fixing microorganism that induces nodulation on several angiosperm plant families and has received a lot of attention due to its role in the nitrogen economy of its hosts (Verma et al., 2009). Several plant-pathogenic taxa include Streptomyces acidiscabies, Streptomyces europaeiscabiei, Streptomyces scabies, and Streptomyces turgidiscabies which cause potato scab (Loria et al., 2006; Bignell et al., 2010); Clavibacter michiganensis with several subspecies and pathogen for alfalfa (C. michiganensis subsp. insidiosus), maize (C. michiganensis subsp. nebraskrensis), potato (C. michiganensis subsp. michiganensis) and wheat (C. michiganensis subsp. tessellarius); Leifsonia xyli subsp. xyli which causes ratoon stunting disease of sugarcane (Young et al., 2006); Curtobacterium flaccumfaciens which affects several Phaseolus and Vigna species, Beta vulgaris species (red and sugar beet), Ilex opaca (American holly), Tulipa species (tulips), and Euphorbia pulcherrima (poinsettia) (Saddler and Messenber-Guimaraes, 2012); Rathayibacter iranicus and Rathayibacter tritici which cause gumming in several grasses and wheat (Evtushenko and Dorofeeva, 2012).
In the last decade, many reports on the isolation and diversity of plant-associated and endophytic actinobacteria from wild plants and crops have been published. In many of these studies, a neutral or a plant growth promotion effect was observed. The isolation and identification of actinobacteria in healthy internal root tissues of wheat was reported by Coombs and Franco (2003a); these authors further demonstrated the colonization of germinating wheat by one of the isolated strains, Streptomyces sp. EN27 (Coombs and Franco, 2003b). A Streptomyces strain, WYEC108, isolated from linseed rhizosphere soil in Great Britain (Crawford et al., 1993) was able to colonize the roots of Pisum sativum, increased the number and size of root nodules, and enhanced the assimilation of iron and other nutrients by the plant (Tokala et al., 2002). Several actinobacterial strains recovered from wild plants adapted to poor soil and severe climate conditions of the Algerian Sahara desert were reported by Goudjal et al. (2013). Some of these strains produced the auxin indol acetic acid (IAA), which promoted seed germination and root elongation when tomato seeds were treated with bacterial supernatants.
The search of endophytic actinobacteria as biological control agents of plant disease is also of interest given their ability to colonize healthy plant tissues and produce antibiotics in situ (Kunoh, 2002; Cao et al., 2004). Maize (Zea mays), an important crop cultivated in many countries, especially in tropical areas, was also screened for the presence of bioactive actinobacteria (de Araújo et al., 2000). Endophytic streptomycetes isolated from healthy banana plants (Musa sp.), were studied for the ability to produce antifungal molecules that inhibited the growth of Fusarium oxysporum, which causes fusarium wilt (Cao et al., 2005). Similarly, Streptomyces strains were isolated from tomato and native plants of the Algerian Sahara and screened for biocontrol activity against Rhizotocnia solani (Goudjal et al., 2014).
Several studies have focused on the diversity and distribution of actinobacterial communities in plants, these works have provided information about the most common taxa found, e.g., the genus Streptomyces, but have also discovered new plant-actinobacteria associations as those represented by the interaction Micromonospora-nitrogen fixing plants.
Members of the genera Microbispora, Micromonospora, Nocardia, Streptosporangium, and Streptoverticillium were recovered from the surface of sterilized roots of different plant species in Italy (Sardi et al., 1992) and of maize in Brazil (de Araújo et al., 2000). Interestingly, the genus Microbispora was the most abundant genus recovered in maize (44%), followed by Streptomyces and Streptosporangium. A diverse collection of 11 native Korean plants were screened for the presence of endophytic actinobacteria. Streptomyces was the most common taxon accounting for almost 50% of the strains isolated and followed by the genera Microbacterium, Microbispora, Micrococcus, Micromonospora, Rhodococcus, and Streptacidiphilus. Single isolates representing the genera Arthrobacter, Dietzia, Herbiconiux, Kitasatospora, Mycobacterium, Nocardia, Rathayibacter, and Tsukamurella were also recovered (Kim et al., 2012).
Kaewkla and Franco (2013) demonstrated the high diversity of actinobacterial strains distributed in native Australian plants using highly designed isolation protocols which included low concentration isolation media, plating larger quantities of plant sample and long incubation times (up to 16 weeks). These authors reported the isolation of >500 actinobacterial strains that were identified in 16 different genera. Again, the genus Streptomyces accounted for >60% of the isolates.
Although the percentage of plant species sampled at present is very low, medicinal plants have received special attention given their importance as potential reservoirs of actinobacterial communities that produce compounds with biotechnological application. Qin et al. (2009, 2012) conducted a thorough study screening medicinal plants growing in the tropical rain forests in Xishuangbanna, China. These authors focused on the isolation of non-streptomycetes and found that the genus Pseudonocardia was the predominant taxon, followed by Nocardiopsis, Micromonospora, and Streptosporangium while almost 25% of the strains could not be identified at the genus level. An in depth analysis of the plant Maytenus austroyunnanensis applying culture- dependent and independent methods revealed an immense diversity reporting genera such as Actinostreptospora, Amnibacterium, Catenuloplanes, Quadrisphaera, and Pseudokineococcus which were previously unknown to reside inside plant tissues (Qin et al., 2012).
A list of endophytic and plant-associated actinobacteria recovered from different plant species and their potential application in agriculture is presented in Table 1.
In recent years, metagenomic analyses have been used to determine the bacterial communities of several agriculturally important crops. These studies have shown that actinobacteria are present in many of these plant microbiomes. Okubo et al. (2014) demonstrated that while the shoots of two field-grown rice cultivars collected in Nipponbare and Kasalath were dominated by Alphaproteobacteria (approximately 52%), the actinobacterial populations made up to 15% of the bacterial community structure. The characterization of the natural microbiome of Vitis vinifera leaves in Portugal reported a high diversity of proteobacteria, firmicutes, and actinobacteria, where the latter group accounted for approximately 19% of the microbial community composition and members of the families Corynebacteriaceae, Microbacteriaceae, and Kineosporiaceae were identified (Pinto et al., 2014).
A recent study to determine the bacterial communities of Olea europaea L. cultivars collected from different regions in the Mediterranean basin also confirmed the presence of actinobacterial populations on the olive leaf endosphere. An interesting conclusion of this work was that soil, climate conditions, and geographical distances had little effect on the endophytic microbial community composition (Müller et al., 2015). In another study, the root microbiota of Lactuca sativa cultivars and its wild ancestor Lactuca serriola were analyzed, the lettuce microbiota was dominated by Proteobacteria and Bacteriodetes, but Chloroflexi and Actinobacteria were also abundant (Cardinale et al., 2015). The composition of the actinobacterial population included members of the families Micromonosporaceae and Nocardioaceae but also the genera Actinoplanes, Aeromicrobium, Arthrobacter, Demequina, and Streptomyces. Interestingly, the domesticated cultivar (L. sativa) was richer in species diversity than its wild counterpart L. serriola. Unfortunately for most of the above studies, the function of these microorganisms on their host plants is unknown. In the case of lettuce, which is one of the raw foods widely consumed, it has been suggested that bacteria present in the plant's root such as Streptomyces, may serve as biological control agents by producing antibiotics to eliminate potential human pathogens (e.g., enterobacteria) (Cardinale et al., 2015).
Several soil microbiomes related to Andropogon gerardii, Schizachyrium scoparium, Lespedeza capitata, and Lupinus perennis grown in communities which varied in plant richness (1–16 species) were determined (Bakker et al., 2014). In this study the antagonistic activity and community structure of Streptomyces populations was assessed in relation to the species plant richness. The authors reported that the diversity and richness of bacterial and Streptomyces communities displayed different relationships with biotic and abiotic soil characteristics, therefore influencing bacterial communities.
The roots, leaves, and stems are the main plant tissues that have been screened for the presence of bacteria, however, nitrogen fixing nodules produced by legumes and actinorhizal plants are also an important reservoir of microorganisms. Nodules are rich in nutrients and therefore can also be colonized by bacteria unrelated to rhizobial or Frankia symbiotic nitrogen fixation.
Actinobacterial strains identified in the genera Agromyces, Curtobacterium, Microbacterium, Micromonospora, and Streptomyces have been reported from nodule tissues (Sturz et al., 1997; Trujillo et al., 2006, 2007, 2010; Zakhia et al., 2006; Muresu et al., 2008; Stajkoviæ et al., 2009; Deng et al., 2011; Hoque et al., 2011; Li et al., 2011; Carro et al., 2012a). Of these, the genera Microbacterium and Micromonospora were the most frequently isolated. Host plants inoculated with some of these strains showed better growth and development in comparison with non-inoculated controls suggesting a beneficial effect (Trujillo et al., 2010, 2014b; Deng et al., 2011; Martínez-Hidalgo et al., 2014). However, our knowledge about these new plant-microbe interactions is still very poor given the limited data currently available.
In light of their ecological importance, Frankia as a provider of nitrogen to actinorhizal plants, and Streptomyces as a plant pathogen for important crops such as potato, these bacteria have been under research for many decades, but this is not the case for most of other reported plant-actinobacteria interactions. However, in the last 10 years the interaction Micromonospora-nitrogen fixing plants is gaining attention due its potential application in downstream biotechnological applications, especially in the area of agriculture. In the following sections we will provide a general overview on the past and present status of Micromonospora and its close interaction with legumes and actinorhizal plants.
Micromonospora and Nitrogen Fixing Nodules: A Universal Plant-Microbe Interaction?
The actinobacterium Micromonospora was first described in 1923 (Ørskov, 1923). The first strains originated from soil and Jensen (1932) pointed out the importance of this microorganism in this niche. This bacterium belongs to the family Micromonosporaceae and includes aerobic, filamentous, spore-producing and mesophilic microorganisms. Micromonospora colonies are usually pigmented and range in color from orange, red, or brown. In many old cultures a brown-black, or black mucous mass of spores is observed. The formation of single spores is the main morphological characteristic of the genus Micromonospora; however, spores are also produced in dense clusters on the surface or completely embedded in the substrate mycelium (Figure 1) (Genilloud, 2012; Trujillo et al., 2014a).
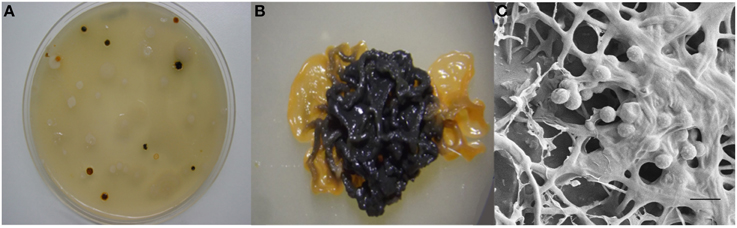
Figure 1. Morphological features of Micromonospora. (A) Micromonosporae isolates recovered from a nitrogen fixing nodule. (B) 14 day old colony producing brown-black spores. (C) Scanning electron micrograph of a mucous mass of spores. Bar, 1 μm (Carro, 2009; Alonso de la Vega, 2010).
The presence of Micromonospora has been reported from many geographical sites worldwide and although soil is the most frequent source of isolation, marine, aquatic sediments and mangrove environments are also inhabited by this microorganism (Maldonado et al., 2009; Genilloud, 2012; Trujillo et al., 2014a). In recent years Micromonosporae have been reported as major components of nitrogen fixing root nodules of both leguminous and actinorhizal plants (Valdés et al., 2005; Trujillo et al., 2006, 2007, 2010; Garcia et al., 2010; Carro et al., 2012a, 2013a). Isolation of Micromonospora strains from internal nodular tissues has been reported from the legumes Arachis hypogaea, Cicer arietinum, Glycine max, Lens culinaris, Lupinus angustifolius, Lupinus gredensis, Medicago sativa, Melilotus sp., Mucuna sp., Ononis sp., Ornithopus sp., Phaseolus sp., Trifolium sp., and Vicia sp. The isolation of Micromonospora strains usually requires selective isolation procedures to favor its slow growth, however, in all the above examples, the same isolation protocol as that used for the isolation of rhizobia was applied (Cerda, 2008; Rodríguez, 2008; Carro, 2009; Alonso de la Vega, 2010; Trujillo et al., 2010).
Actinorhizal plants that have been sampled to date in Mexico, Spain, Canada, and France include the species Alnus viridis, Casuarina equisetifolia, Coriaria myrtifolia, Elaeagnus x ebbingei, Hippophae rhamnoides, Myrica gale, and Morella pensylvanica (Valdés et al., 2005; Trujillo et al., 2006; Carro et al., 2013a). Except for the study of Valdés et al. (2005), the isolation of Micromonospora from actinorhizal nodules also followed the same isolation protocols as that of legumes, using yeast-mannitol agar as isolation medium (Vincent, 1970). Currently our group maintains a collection of ~2000 isolates recovered from diverse legume and actinorhizal plants species collected in Spain, France, Germany, Ecuador, Nicaragua, and Australia but our hypothesis is that Micromonospora is also present in those plant species which have not been sampled to date. In the case of legumes, the above examples indicate how Micromonospora had been largely overlooked in this niche due to its slow growth as compared to rhizobial strains which can be readily recovered from isolation plates after 3–5 days while Micromonospora strains usually appear after 7–10 days on the same plates. While the work carried by Carro et al. (2013a) strongly suggests that this microorganism is also a normal occupant of actinorhizal nodules. Thus, the systematic recovery of Micromonospora populations strongly suggests that this bacterium closely interacts with the host plant and nitrogen-fixing bacteria occupying the same niche.
The biogeographical and species distribution of Micromonosporae isolated from nitrogen fixing nodules of legumes and actinorhizal plants sampled hitherto is presented in Table 2.
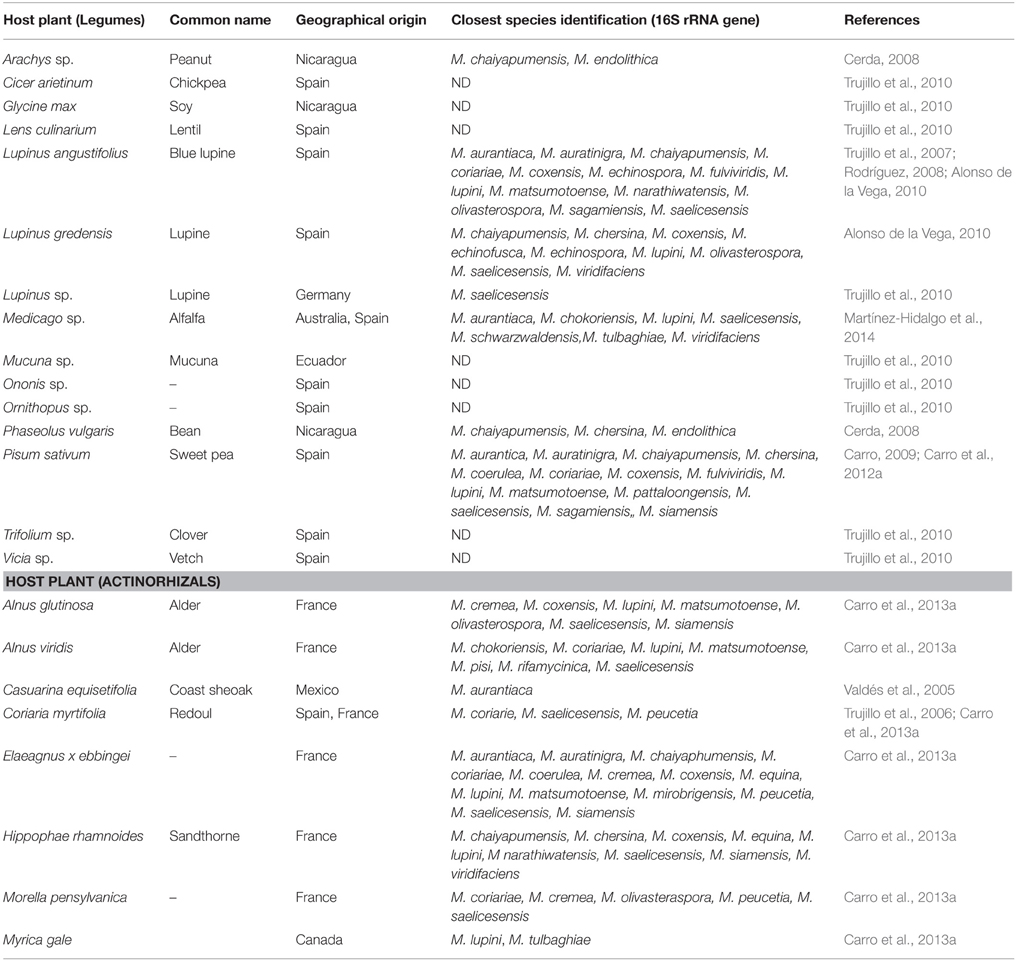
Table 2. Biogeographical and species distribution of Micromonosporae in nitrogen fixing nodules of legumes and actinorhizal plants sampled.
Distribution, Localization and Genetic Diversity of Micromonospora in Nitrogen Fixing Nodules
The distribution of Micromonospora strains in the nitrogen fixing nodules sampled so far indicate that its distribution is not homogeneous and it varies from nodule to nodule and plant to plant (Trujillo et al., 2010; Carro et al., 2012a).
The distribution pattern of Micromonospora in Lupinus spp. is highly variable with no isolates for some nodules to as many as approximately 30 (Alonso de la Vega, 2010; Trujillo et al., 2010). Variation is also reported from plant to plant and from different nodules of the same plant (Trujillo et al., 2010). A comparison of the species Lupinus angustifolius and Lupinus gredensis collected in the same geographical area in Spain, indicated that 67 and 60% of the plant samples screened (17 in total) contained the target microorganism, respectively. Out of the 45 nodules chosen for isolation, 95 Micromonospora strains were recovered, 74 from L. angustifolius and 21 from L. gredensis. Interestingly, 48% of the nodules did not appear to contain any Micromonospora strains (Alonso de la Vega, 2010).
In terms of the bacterial species distribution, Micromonospora saelicesensis and Micromonospora lupini were the most abundant, nevertheless the diversity determined on the basis of 16S rRNA gene sequencing was very high (Alonso de la Vega, 2010; Trujillo et al., 2010). These authors also screened lupine plants at different growth stages which corresponded to young, maximum growth, and flowering plants. In this case, the number of bacteria increased in parallel to the plant growth and decreased as the plants became old.
As for the legume Pisum sativum, a similar pattern of distribution was observed. However, for this plant, at least one Micromonospora strain was recovered from every nodule sampled (Carro et al., 2012a). It is also important to note that while lupine plants were collected in the field, all Pisum sativum samples originated from cultivation fields where chemical fertilizers are applied periodically (Carro et al., 2012a).
In a recent study, Carro et al. (2013a) screened several actinorhizal plants and recorded the number of Micromonospora strains and species found. Micromonospora strains were recovered from all plants sampled, and, as in the case of legumes, the number of isolates also varied significantly. High numbers of Micromonospora strains were isolated from Alnus, Elaeagnus, and Hippophae nodules, while the number of isolates was much lower in Myrica, Morella, and Coriaria nodules. Similarly to legumes, most isolates were related to M. saelicesensis and M. lupini but M. coriariae was also isolated in high numbers. The latter species was first reported from Coriaria myrtifolia nodules (Trujillo et al., 2006).
The first Micromonospora strains isolated from nitrogen fixing nodules were considered contaminants because it was assumed that the spores produced by this microorganism were soil contaminants that had resisted the sterilization protocols. However, the absence of other fast-growing sporulating microorganisms, e.g., fungi or Streptomyces strongly indicated that the strains had originated from the internal plant tissues (Trujillo et al., 2010). Applying fluorescent in situ hybridization (FISH) and transmission electronic microscopy (TEM), Micromonospora lupini Lupac 08 was localized inside the nodular tissues of lupin suggesting a close interaction between the host plant and the bacterium (Rodríguez, 2008; Trujillo et al., 2010). Further experiments using a Micromonospora strain tagged with green fluorescent protein to trace the microorganism in planta are in the process of completion.
The degree of genetic variation of Micromonospora strains recovered from the nitrogen-fixing nodules of various plants was analyzed using several molecular typing techniques (e.g., BOX–PCR, ARDRA, RFLP, RAPDS) (Cerda, 2008; Carro, 2009; Alonso de la Vega, 2010; Trujillo et al., 2010; Carro et al., 2012a; Martínez-Hidalgo et al., 2014). Highly diverse genetic fingerprint profiles were found among the isolates studied, indicating that they were not clones; the diversity found was unexpectedly high considering that in some cases, the strains analyzed were isolated from the same nodule (Alonso de la Vega, 2010). Subsequently, taxonomic studies carried for some of these isolates confirmed that many of these bacterial strains represented new species and include Micromonospora coriariae (Trujillo et al., 2006); Micromonospora lupini and Micromonospora saelicesensis (Trujillo et al., 2007); Micromonospora pisi (Garcia et al., 2010); Micromonospora cremea, Micromonospora zamorensis, and Micromonospora halotolerans (Figure 2). The latter three strains were isolated from the rhizospheric soil of the sampled plants (Carro et al., 2012b, 2013b).
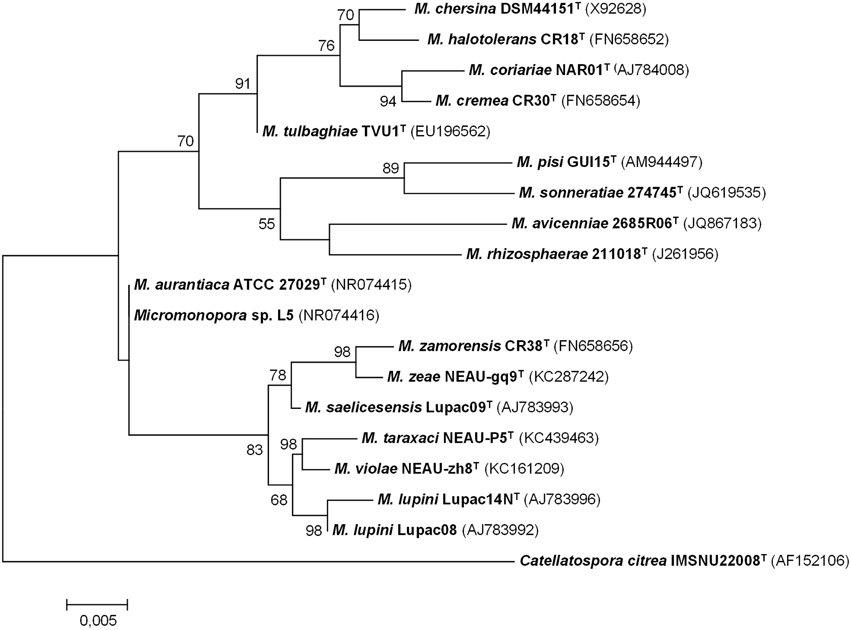
Figure 2. Maximum-likelihood phylogenetic tree based on 16S rRNA gene sequences of Micromonospora species isolated from plant material and rhizospheric soil. There were 1408 nucleotides in the final dataset. Analyses were carried in MEGA 6 software. Bar indicates 0.005 substitutions per nucleotide position (Based on references provided in Table 2).
The species M. saelicesensis is the most frequently isolated from the nodule tissues in both legume and actinorhizal plants, followed by the species M. lupini (Cerda, 2008; Carro, 2009; Alonso de la Vega, 2010; Trujillo et al., 2010; Carro et al., 2012a). Furthermore, the number of new species found in this niche also appears to be very high as commented above. To expand the taxonomic studies of the genus Micromonospora, Carro et al. (2012a) carried out a multilocus sequence analysis study based on five loci and over 90 Micromonospora isolates recovered from the rhizosphere and plant tissues (nodules) of P. sativum. These studies were complemented with DNA-DNA hybridization analyses to confirm the high diversity at the species level (Carro et al., 2012a) and revealed that many of the new isolates represent new species (Carro et al., 2012b, 2013b).
Genome Features of Micromonospora Isolated from Nodules
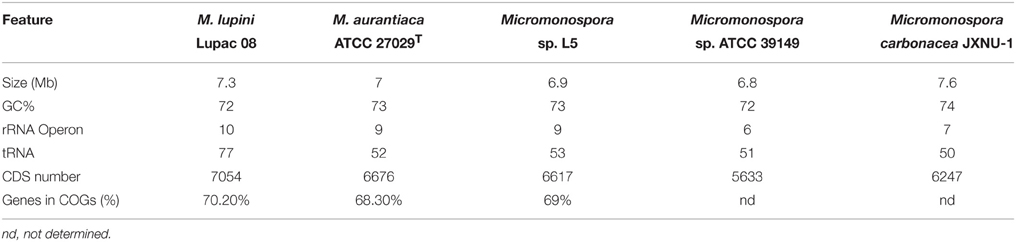
Very few Micromonospora strains have been sequenced. At present, only five Micromonospora genomes are available in the public databases: Micromonospora sp. strain L5 and M. lupini Lupac 08 and isolated from nodules of Casuarina equisetifolia and Lupinus angustifolius, respectively (Alonso-Vega et al., 2012; Hirsch et al., 2013). The remaining are the soil isolates Micromonospora aurantiaca ATCC 27029T (Hirsch et al., 2013), Micromonospora sp. ATCC 39149 (Accession No. GCF_000158815.1) and Micromonospora carbonacea JXNU-1 (Jiang et al., 2015). Several genomic characteristics of the strains are presented in Table 3. Actinobacterial genomes are usually larger than those of most other bacteria, e.g., proteobacteria and Micromonospora is no exception, the currently available genomes range from 6.9 to 7.3 Mb and share a similar GC content (72–74%).
The genome sequence of strain Lupac 08 was determined to identify genomic traits potentially involved in this plant-microbe interaction (Alonso-Vega et al., 2012; Trujillo et al., 2014b). The annotated genome disclosed various traits potentially involved in the capacity of this bacterium to alternate a lifestyle as a saprophyte in the soil and as an endophyte inside the root nodules (Trujillo et al., 2014b). The genome of strain Lupac 08 has a circular chromosome of 7.3 Mb with a GC content of 71.9% and lacking plasmids. A total of 10 rRNA genes were identified, specifically 3 5S rRNA, 4 16S rRNA, and 3 23S rRNA genes. In addition 77 tRNA genes were predicted (Alonso-Vega et al., 2012). Approximately, 62% (4338 CDSs) of the genes were assigned a biological function while 38% were annotated hypothetical open reading frames with unknown biological activities (Alonso-Vega et al., 2012). The genome of Micromonospora sp. L5 is smaller, 6.9 Mb, a GC content of 72.9% and 6332 open reading frames (Hirsch et al., 2013). This strain is highly related to M. aurantiaca ATCC 27029T and average nucleotide identity values (ANI) of their genomes strongly suggest that Micromonospora sp. L5 belongs to this species. The number of tRNAs identified in Micromonospora sp. L5 is 52 (Hirsch et al., 2013) which is much lower when compared to the 77 tRNAs identified in M. lupini 08. Indeed, the latter strain has one of the largest numbers of tRNAs reported for actinobacteria sequenced to date. The number of rRNA and tRNA genes in a genome appear to be correlated and is an indication of positive selection related to the time of response of a bacterium to adapt to its environment (Dethlefsen and Schmidt, 2007; Yano et al., 2013).
The core genome of the strains M. lupini Lupac 08, M. aurantiaca ATCC 27029T and Micromonospora sp. L5 was determined and the results indicated that the strains shared a common gene pool of only approximately 32% suggesting a high degree of genomic diversity (Trujillo et al., 2014b). As expected, the strains M. aurantiaca and Micromonospora L5 with 85% genome similarity confirm their close relationship. M. lupini on the other hand appears to be very different, with 66.6% of its genome being strain specific. As more Micromonospora genomes are sequenced the core genome should be better defined.
A number of genomic traits that probably participate in the plant/soil life style of endophytic Micromonospora include transport and secretion systems. Several genes coding for transport and secretion systems which may be involved in plant colonization were also identified. The number of transporters is slightly higher in M. lupini Lupac 08 than in Micromonospora L5, and included ATP dependent (mainly of the ABC family type), ion channels, PTS (phosphotransferase) and secondary transporters (Trujillo et al., 2014b).
Micromonospora Lupini Lupac 08: A Friendly Bacterium Highly Equipped with Plant Cell Wall Degrading Enzymes
Micromonosporae are well-known for their capacity to produce high numbers of cellulases, these enzymes very likely contribute to the turn-over of decayed material in different habitats (de Menezes et al., 2008, 2012). However, the presence of high numbers of these molecules and other plant-cell wall degrading enzymes in beneficial endophytic bacteria is usually very low (Krause et al., 2007; Mastronunzio et al., 2008; Taghavi et al., 2010; Pujic et al., 2012).
The genome of strain Lupac 08 contains a high number of genes encoding enzymes potentially involved in plant cell wall degradation. Approximately 10% of the genome codes for carbohydrate metabolism, and almost 200 out of the 685 genes have a putative hydrolytic function. Hydrolytic activities for cellulose, pectin, starch, and xylan, were confirmed in the laboratory and indicate that this strain could degrade plant cell wall components in a way similar to that of phytopathogen bacteria (Trujillo et al., 2014b). Plant-polymer degrading enzymes are thought to be involved in internal plant colonization (Compant et al., 2005). Plant pathogenic fungi and bacteria usually enter plant tissues by degrading plant cell wall components using several hydrolases which include cellulases and endoglucanases. On the other hand, genome data show that non-pathogenic (endophytic or symbiotic) microorganisms contain a low set of plant-polymer degrading enzymes (Krause et al., 2007; Mastronunzio et al., 2008; Taghavi et al., 2010). In the case of M lupini, the genome of this microorganism revealed a high number of hydrolytic enzymes (e.g., cellulases, xylanases, endoglucanases) with the potential to degrade plant tissues (Figure 3). However, green-house experiments show that when host plants are inoculated with strain Lupac 08 no damage is produced. On the contrary, M. lupini stimulates nodulation and plant growth (Cerda, 2008; Trujillo et al., 2014b). Therefore, if the plant does appear to be negatively affected by these enzymes, what is their potential function when the bacterium interacts with its host plant? Our group is currently working on this subject, some of the loci, especially those related to cellulose metabolism may participate in other processes such as cellulose biosynthesis (Robledo et al., 2008, 2012; Mba Medie et al., 2012). Several genes coding for plant cell-wall degrading enzymes were also located in the genome of Micromonospora sp. L5 (Hirsch et al., 2013). Similarly to strain Lupac 08, target substrates include cellulose, hemicellulose, pectin, starch, and xylan, however, the number of loci involved in carbohydrate transport and metabolism are slightly lower in strain L5 (8.9%), as compared to strain Lupac 08 (9.7%) (Trujillo et al., 2014b).
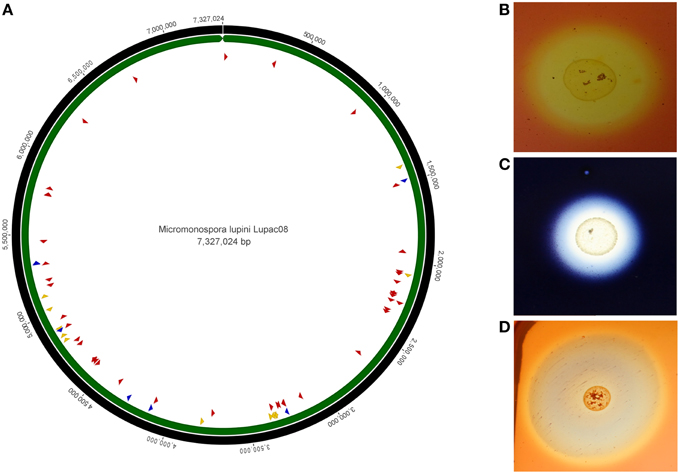
Figure 3. Circular genome representation of Micromonospora lupini, Lupac 08. (A) Distribution of various plant-cell wall hydrolytic enzyme loci. Red, cellulases, and cellulose-binding sites; blue, pectinases; yellow, xylanases. (B) In vitro cellulase degradation. (C) In vitro starch degradation. (D) In vitro xylanase degradation (Based on Trujillo et al., 2014b).
Bacterial endophytic colonization is still a poorly understood process, in part because it is very complex. For microorganisms that colonize the roots, plant exudates appear to play a crucial role (Badri et al., 2009). Molecules present in root exudates may serve as carbon sources for microorganisms and therefore, these are attracted to the plant roots (Shidore et al., 2012). Thus, plant exudates may act as signals that influence the ability of a bacterium to colonize the root or survive in the rhizosphere. These signals may induce the alteration of specific gene expression patterns in the bacterium, which in turn may influence its interaction with the plant (Morrissey et al., 2004; Mark et al., 2005; Shidore et al., 2012). While it is considered that plant exudates affect the behavior of rhizospheric microorganisms, our knowledge as to how these molecules influence bacterial gene expression is still very limited (Mark et al., 2005). Furthermore, it is not known how these altered bacterial genes affect the plant-microbe interaction process and only a few studies are available (Morrissey et al., 2004; Mark et al., 2005; Shidore et al., 2012).
In the case of the Micromonospora-plant interaction, it could be that the plant's root exudates might be involved in the repression of hydrolytic enzyme genes (e.g., cellulases, xylanases, etc.) from the bacterium which, if expressed during its interaction with the plant would be detrimental upon infection. The effect on Azoarcus sp. gene expression upon exposure to plant root exudates was recently reported (Shidore et al., 2012). This study concluded that the genes expressed by Azoarcus strain BH72 upon exposure to the plant's root exudates influenced the colonization of the roots (Shidore et al., 2012). In this sense, the genome of M. lupini contains many regulatory genes located near plant cell wall degrading loci suggesting that these genes are under strong regulation, which in turn, may be directly related to the surrounding environment, soil, or plant tissues (Trujillo et al., 2014b).
Micromonospora, A Plant Growth Promoter with Wide Application in Agriculture
Plant growth promoting bacteria (PGPB) are defined as soil bacteria that facilitate plant growth and are often found in association with plant roots, leaves, flowers, or within plant tissues. Many of these bacteria are found in the plant rhizoplane and rhizosphere but other are endophytic and able to colonize the internal plant tissues (Glick, 2015). Plant growth promoting bacteria have been reported to positively affect plants in a number of ways, directly by facilitating resource acquisition (e.g., nitrogen fixation, phosphorous, iron) or controlling plant hormone levels, or indirectly by lowering the inhibitory effects of plant pathogen microorganisms (e.g., biocontrol agents).
The current data about the interaction of Micromonospora with legume and actinorhizal plants is limited, and therefore the bacterium's ecological role inside the roots nodules and its interaction with the nitrogen fixing bacteria (rhizobia/Frankia) is unknown. Plant co-inoculation studies indicate that Micromonospora acts as a plant growth promoting bacterium with a positive effect on the plant (Martínez-Hidalgo et al., 2014; Trujillo et al., 2014b). Nodulation and nitrogen tests were carried out on Lupinus and Phaseolus, these studies showed that Micromonospora is not able to induce nodules or fix nitrogen but a positive effect on the growth of the plant was observed by an increase in the number of nodules and the height of the plants which had been inoculated with both microorganisms when compared to the plants treated with only one of the two strains (Cerda, 2008). Furthermore, when Micromonospora and the nitrogen-fixing bacterium (Bradyrhizobium or Rhizobium, respectively) were grown together, they were compatible and did not inhibit the growth of each other. Interestingly, Micromonospora did inhibit the growth of several Frankia strains; furthermore the latter strains came from different plant species (Carro et al., 2013a). On the other hand no inhibition was observed between Micromonospora and Frankia when the strains originated from the same plant (Carro et al., 2013a).
Studies carried out with Trifolium plants yielded similar results. Micromonospora lupini Lupac 08 stimulated plant growth when it was co-inoculated with Rhizobium sp. on clover plantlets and these were grown in a greenhouse (Trujillo et al., 2014b). In general, the number of nitrogen-fixing nodules increased in plants treated with both bacteria as compared to the plants inoculated only with the Rhizobium strain. Overall, the plants inoculated with both bacteria exhibited better growth and increased shoot length compared to single-strain treatments (Trujillo et al., 2014b).
Solans (2007) studied the plant promotion effect of three actinobacterial strains isolated from the plant species Discaria trinervis which included a Micromonospora strain. The inoculation experiments of D. trinervis grown in glass tubes with vermiculite-sand was done using pure mycelia suspensions and/or supernatants obtained from the actinobacterial cultures grown for 8 days. Plants inoculated with mycelium plus supernatant from Micromonospora strain BCRU-MM18 had a higher shoot length than the control plants and it was proposed that this effect was probably due to the presence of several plant hormones such as zeatin, IAA, and gibberellic acid. Further studies confirmed that strain BCRU-MM18 produced significant amounts of IAA (9.03 ng/ml), giberellic acid (9.03 ng/ml), and zeatin (270 μg/ml); in all cases these amounts were higher than those produced by the nitrogen fixer Frankia sp. BCU110501 (Solans et al., 2011). The same Micromonospora strain (BCRU-MM18) was co-inoculated in Medicago sativa which had also been inoculated with the nitrogen fixer Sinorhizobium meliloti in the presence of high nitrogen content. Unexpectedly, a promotion of nodulation was observed despite the high amounts of nitrogen present (7 mM) which usually inhibit nodulation (Solans et al., 2009). The above studies showed the positive effect that Micromonospora had on the symbiosis of both leguminous and actinorhizal plants, especially in increasing nodulation rates.
Recently, Micromonospora strains isolated from wild alfalfa plants collected in several sites in Spain were studied for their plant growth and nutrient content effect on this legume. Selected strains significantly increased the nodulation of Medicago sp. inoculated with Ensifer meliloti and also the plant's efficiency for nitrogen uptake. Furthermore, aerial growth, shoot-to-root ratio and increase in levels of key nutrients was also reported (Martínez-Hidalgo et al., 2014). These authors also discussed the importance of choosing the most effective strains.
The wide distribution of Micromonospora among nitrogen fixing plants (both legumes and actinorhizals) differs from that of rhizobia or Frankia which are limited to a narrow host range of legumes and angiosperms, respectively. The capacity of infection by Micromonospora with a positive effect for its host plant may be regarded as an advantage for downstream biotechnological applications and the potential to use this bacterium as a plant growth promoter in combination with rhizobia or Frankia.
The Micromonospora Metabolome and its Potential Role in Plant-microbe Communication Signals
Microbial secondary metabolites have been the subject of many research projects, mainly with the aim to discover new compounds with biotechnological application (Miao and Davies, 2010; Genilloud, 2014). However, our knowledge about the ecological role of these compounds is very limited. It is proposed, that in the environment, these natural products serve as allelochemicals and signaling molecules to communicate with organisms, in this case, with the plant (Badri et al., 2009). Udwary et al. (2011) recently reported the identification of several biosynthetic gene clusters coding for secondary metabolites in the genome of Frankia. In this work, it was proposed that some of these compounds could function as communication molecules to establish the symbiotic interaction between Frankia and the host plant (Udwary et al., 2011). The potential role of lectins produced by Frankia alni ACN14a to permit binding of the bacterial cells to the roots of the host plant was suggested by Pujic et al. (2012). In another study, a hybrid (PKS)/NRPS protein produced by Trichoderma virens was proposed to induce the defense mechanisms of maize (Mukherjee et al., 2012).
Moreover, Conn et al. (2008) demonstrated that culture filtrates obtained from Micromonospora sp. strain EN43 isolated from healthy wheat tissues were able to induce several plant defense systems in Arabidopsis thaliana. When the bacterium was grown in a minimal medium, the culture filtrate applied to the plant induced the systemic acquired system pathway; however, when grown in a complex medium, the jasmonic acid/ethylene pathway was activated (Conn et al., 2008). Based on these results, the authors suggested that different metabolites were produced under the two conditions tested and that these compounds were responsible for the activation of the different defense mechanisms in the plant (Conn et al., 2008). In addition, it was also proposed that a physical contact of the bacterium and the plant may be required for the defense mechanisms to be activated. Overall, the above examples show the potential ecological role of secondary metabolites in plant-microbe interactions.
The information derived from sequenced actinobacterial genomes have revealed that these microorganisms have the biosynthetic potential to make far more natural products than was realized before genome sequences were available (Genilloud, 2014). Only a small fraction of endophytic bacteria have been characterized and they remain as an untapped resource of novel bioactive small molecules (Qin et al., 2011; Brader et al., 2014). As mentioned above, some of these metabolites are speculated to affect the physiological conditions of host plants including growth and disease resistance (Conn et al., 2008; Udwary et al., 2011). Micromonosporae strains are also a good source for obtaining natural products (Weinstein et al., 1963; Thawai et al., 2004; Antal et al., 2005; Anzai et al., 2010; Kyeremeth et al., 2014). In this sense, the model strain Micromonospora lupini Lupac 08 is no exception and a family of new anthraquinone molecules with antitumoral activity were isolated and identified (Igarashi et al., 2007, 2011). Moreover, 15 clusters involved in the biosynthesis of secondary metabolites were identified in the genome of M. lupini Lupac 08. These included siderophores, terpenes, butyrolactones, polyketides (PKS), non-ribosomal peptides (NRPS), chalcone synthases and bacteriocins. Approximately 7.4% of the genome was related to genes coding for secondary metabolites.
The production of siderophores by endophytic bacteria is suggested to promote plant growth by sequestering iron from the environment and providing the nutrient to the plant. Alternatively, plant growth promoting bacteria can protect plants by binding the available iron surrounding the roots and limiting access to the nutrient by phytopathogen microorganisms (Glick, 2015). Recently it was shown that a siderophore-producing endophytic streptomyces strain significantly increased root and shoot biomass as compared to a siderophore deficient mutant strain (Rungin et al., 2012). Furthermore, Misk and Franco (2011) reported the capacity of several endophytic siderophore producing Streptomyces strains to suppress root rot in chickpea produced by Phytophtora. In this case, the streptomycete strains were isolated from several legumes. Several gene loci related with the synthesis of siderophores were identified in the genome of M. lupini Lupac 08 and the strain was shown to produce these molecules in the laboratory (Trujillo et al., 2014b). Siderophores produced by Micromonospora may also contribute to the increased root and shoot biomass observed when host plants are inoculated with this bacterium (Martínez-Hidalgo et al., 2014; Trujillo et al., 2014b).
The characterization and identification of secondary metabolites produced by Micromonospora strains isolated from nitrogen fixing plants is at present reduced to three anthraquinones, lupinacidins A, B, and C (Igarashi et al., 2007, 2011). However, the genome of strain Lupac 08 revealed that other metabolites are potentially produced (e.g., terpenes, butyrolactones, polyketides, non-ribosomal peptides etc.). These compounds may act as communication molecules between the microorganism and the plant to allow bacterial colonization (Udwary et al., 2011). Alternatively, as suggested by other studies these metabolites may provide protection against pathogens, either by producing specific control agents or by activating plant defense systems (Conn et al., 2008). Furthermore, some metabolites may be necessary for nutrient uptake (Barry and Challis, 2009; Rungin et al., 2012) All these areas remain to be studied in the interaction Micromonospora-nitrogen fixing plants.
Concluding Remarks
Our knowledge of the interaction between Micromonospora with legumes and actinorhizal plants is in its infancy and a lot more work is required to fully understand this ecological process. Apart from the studies presented above, there is no other information regarding the molecular interaction between Micromonospora and its host plants and how it interacts with other bacteria present in the nitrogen fixing nodules. The current data is promising as it strongly suggests that Micromonospora provides a benefit to the plant. The genome of strain Lupac 08 revealed many features that make this microorganism an excellent candidate as a plant-growth promoter which could be applied to a large number of agriculturally important crops.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to acknowledge past and present members of the laboratory who contributed to some of the studies cited in this work. MT received financial support from the Spanish Ministerio de Economía y Competitividad under project CGL2014-52735-P.
References
Alonso de la Vega, P. (2010). Distribución, Caracterización e Importancia Ecológica de Micromonospora en Nódulos Fijadores de Nitrógeno de Lupinus. Ph.D. thesis, Universidad de Salamanca, Salamanca.
Alonso-Vega, P., Normand, P., Bacigalupe, R., Pujic, P., Lajus, A., Vallenet, D., et al. (2012). Genome sequence of Micromonospora lupini Lupac 08, isolated from root nodules of Lupinus angustifolius. J. Bacteriol. 194, 4135. doi: 10.1128/JB.00628-12
Antal, N., Fiedler, H. P., Stackebrandt, E., Beil, W., Ströch, K., and Zeeck, A. (2005). Retymicin, galtamycin B, saquayamycin Z and ribofuranosyllumchrome, novel secondary metabolites from Micromonospora sp. Tü 6368. I. Taxonomy, fermentation, isolation and biological activities. J. Antibiot. 58, 95–102. doi: 10.1038/ja.2005.12
Anzai, Y., Sakai, A., Li, W., Iizaka, Y., Koike, K., Kinoshita, K., et al. (2010). Isolation and characterization of 23-O-mycinosyl-20-dihydro-rosamicin: a new rosamicin analogue derived from engineered Micromonospora rosaria. J. Antibiot. 63, 325–328. doi: 10.1038/ja.2010.38
Badri, D. V., Weir, T. L., van der Lelie, D., and Vivanco, J. M. (2009). Rhizosphere chemical dialogues: plant-microbe interactions. Curr. Opin. Biotechnol. 20, 642–650. doi: 10.1016/j.copbio.2009.09.014
Bakker, M. G., Schlatter, D. C., Otto-Hanson, L., and Kinkel, L. L. (2014). Diffuse symbioses: roles of plant-plant, plant-microbe and microbe-microbe interactions in structuring the soil microbiome. Mol. Ecol. 23, 1571–1583. doi: 10.1111/mec.12571
Barry, S. M., and Challis, G. L. (2009). Recent advances in siderophore biosyntehsis. Curr. Opin. Biotechnol. 13, 205–215. doi: 10.1016/j.cbpa.2009.03.008
Berendsen, R. L., Pieterse, C. M. J., and Bakker, P. A. H. M. (2012). The rhizosphere microbiome and plant health. Trends Plant Sci. 17, 478–486. doi: 10.1016/j.tplants.2012.04.001
Bignell, D. R., Huguet-Tapia, J. C., Joshi, M. V., Pettis, G. S., and Loria, R. (2010). What does it take to be a pathogen: genomic insights from Streptomyces species. Antonie Van Leeuwenhoek 98, 179–194. doi: 10.1007/s10482-010-9429-1
Bouizgarne, B., and Ben Aouamar, A. A. (2014). “Diversity of plant associated actinobacteria” in Bacterial Diversity in Sustainable Agriculture, ed D. K. Maheswari (Cham: Springer), 41–99.
Brader, G., Compant, S., Mitter, B., Trognitz, F., and Sessitsch, A. (2014). Metabolic potential of endophytic bacteria. Curr. Opin. Biotechnol. 27, 30–37. doi: 10.1016/j.copbio.2013.09.012
Callaham, D., Deltredici, P., and Torrey, J. G. (1978). Isolation and cultivation in vitro of the actinomycete causing root nodulation in Comptonia. Science 199, 899–902. doi: 10.1126/science.199.4331.899
Cao, L., Qiu, Z., You, J., Tan, H., and Zhou, S. (2004). Isolation and characterization of endophytic Streptomyces strains from surface-sterilized tomato (Lycopersicon esculentum) roots. Lett. Appl. Microbiol. 39, 425–430. doi: 10.1111/j.1472-765X.2004.01606.x
Cao, L., Qiu, Z., You, J., Tan, H., and Zhou, S. (2005). Isolation and characterization of endophytic streptomycete antagonists of fusarium wilt pathogen from surface-sterilized banana roots. FEMS Microbiol. Lett. 247, 147–152. doi: 10.1016/j.femsle.2005.05.006
Cardinale, M., Grube, M., Erlacher, A., Quehenberger, J., and Berg, G. (2015). Bacterial networks and co-occurrence relationships in the lettuce root microbiota. Environ. Microbiol. 17, 239–252. doi: 10.1111/1462-2920.12686
Carro, L. (2009). Avances en la Sistemática del Género Micromonospora: Estudio de Cepas aisladas de la Rizosfera y Nódulos de Pisum Sativum. Ph. D. thesis, Universidad de Salamanca, Salamanca.
Carro, L., Pujic, P., Trujillo, M. E., and Normand, P. (2013a). Micromonospora is a normal inhabitant of actinorhizal nodules. J. Biosci. 38, 685–693. doi: 10.1007/s12038-013-9359-y
Carro, L., Pukall, R., Spröer, C., Kroppenstedt, R. M., and Trujillo, M. E. (2012b). Micromonospora cremea sp. nov. and Micromonospora zamorensis sp. nov., isolated from the rhizosphere of Pisum sativum. Int. J. Syst. Evol. Microbiol. 62, 2971–2977. doi: 10.1099/ijs.0.038695-0
Carro, L., Pukall, R., Spröer, C., Kroppenstedt, R. M., and Trujillo, M. E. (2013b). Micromonospora halotolerans sp. nov. isolated from the rhizosphere of a Pisum sativum plant. Antonie Van Leewenhoek 103, 1245–1254. doi: 10.1007/s10
Carro, L., Spröer, C., Alonso, P., and Trujillo, M. E. (2012a). Diversity of Micromonospora strains isolated from nitrogen fixing nodules and rhizosphere of Pisum sativum analyzed by multilocus sequence analysis. Syst. Appl. Microbiol. 35, 73–80. doi: 10.1016/j.syapm.2011.11.003
Cerda, E. (2008). Aislamiento de Micromonospora de Nódulos de Leguminosas Tropicales y Análisis de Su Interés Como Promotor del Crecimiento Vegetal. Ph.D. thesis, Universidad de Salamanca, Salamanca.
Compant, S., Duffy, B., Nowak, J., Clément, C., and Barka, E. A. (2005). Use of plantgrowth- promoting bacteria for biocontrol of plant diseases: principles, mechanisms of action, and future prospects. Appl. Environ. Microbiol. 71, 4951–4959. doi: 10.1128/AEM.71.9.4951-4959.2005
Conn, V. M., Walker, A. R., and Franco, M. M. (2008). Endophytic actinobacteria induce defense pathways in Arabidopsis thaliana. Mol. Plant Microb. Interact. 21, 208–218. doi: 10.1094/MPMI-21-2-0208
Coombs, J. C., and Franco, C. M. M. (2003a). Isolation and identification of actinobacteria from surface-sterilized wheat roots. Appl. Environ. Microbiol. 69, 5603–5608. doi: 10.1128/AEM.69.9.5603-5608.2003
Coombs, J. C., and Franco, C. M. M. (2003b). Visualization of an endophytic Streptomyces species in wheat seed. Appl. Environ. Microbiol. 69, 4260–4262. doi: 10.1128/AEM.69.7.4260-4262.2003
Crawford, D. L., Lynch, J. M., Whipps, J. M., and Ousley, M. A. (1993). Isolation and characterization of actinomycete antagonists of a fungal root pathogen. Appl. Environ. Microbiol. 59, 3899–3905.
de Araújo, J. M., Silva, A. C., and Azevedo, J. L. (2000). Isolation of endophytic actinomycetes from roots and leaves of maize (Zea mays L.). Br. Arch. Biol. Technol. 43, 447–451. doi: 10.1590/s1516-89132000000400016
de Menezes, A. B., Lockhart, R. J., Cox, M. J., Allison, H. E., and McCarthy, A. J. (2008). Cellulose degradation by Micromonosporas recovered from freshwater lakes and classification of these actinomycetes by DNA gyrase B gene sequencing. Appl. Environ. Microbiol. 74, 7080–7084. doi: 10.1128/AEM.01092-08
de Menezes, A. B., McDonald, J. E., Allison, H. E., and McCarthy, A. J. (2012). Importance of Micromonospora spp. as colonizers of cellulose in freshwater lakes as demonstrated by quantitative reverse transcriptase PCR of 16S rRNA. Appl. Environ. Microbiol. 78, 3495–3499. doi: 10.1128/AEM.07314-11
Deng, Z. S., Zhao, L. F., Kong, Z. Y., Yang, W. Q., Lindström, K., Wang, E. T., et al. (2011). Diversity of endophytic bacteria within nodules of the Sphaerophysa salsula in different regions of Loess Plateau in China. FEMS Microbiol. Ecol. 76, 463–475. doi: 10.1111/j.1574-6941.2011.01063.x
Dethlefsen, L., and Schmidt, T. M. (2007). Performance of the translational apparatus varies with the ecological strategies of bacteria. J. Bacteriol. 189, 3237–3245. doi: 10.1128/JB.01686-06
Dudeja, S. S., Giri, R., Saini, R., Suneja-Madan, P., and Kothe, E. (2012). Interaction of endophytic microbes with legumes. J. Basic Microbiol. 52, 248–260. doi: 10.1002/jobm.201100063
El-Shatoury, S. A., El-Kraly, O. A., Trujillo, M. E., El-Kazzaz, W. M., El-Sayeda, G. E.-D., and Dewedar, A. (2013). Generic and functional diversity in endophytic actinomycetes from wild Compositae plant species at South Sinai - Egypt. Res. Microbiol. 164, 761–769. doi: 10.1016/j.resmic.2013.03.004
El-Tarabily, K. A., Nassar, A. H., Hardy, G. E., and Sivasithamparam, K. (2009). Plant growth promotion and biological control of Pythium aphanidermatum, a pathogen of cucumber, by endophytic actinomycetes. J. Appl. Microbiol. 106, 13–26. doi: 10.1111/j.1365-2672.2008.03926.x
Evtushenko, L. I., and Dorofeeva, L. V. (2012). “Genus Rathayibacter,” in Bergey's Manual of Systematic Bacteriology, Vol. 5, eds M. Goodfellow, P. Kämpfer, H.-J. Busse, M. E. Trujillo, K.-I. Suzuki, W Ludwig, et al. (New York, NY: Springer), 949–960.
Francis, S., Holsters, M., and Vereecke, D. (2010). The Gram positive side of plant-microbe interactions. Environ. Microbiol. 12, 1–12. doi: 10.1111/j.1462-2920.2009.01989.x
Garcia, L. C., Martínez-Molina, E., and Trujillo, M. E. (2010). Micromonospora pisi sp. nov., isolated from root nodules of Pisum sativum. Int. J. Syst. Evol. Microbiol. 60, 331–337. doi: 10.1099/ijs.0.012708-0
Genilloud, O. (2012). “Genus Micromonospora,” in Bergey's Manual of Systematic Bacteriology, Vol. 5, eds M. Goodfellow, P. Kämpfer, H.-J. Busse, M. E. Trujillo, K.-I. Suzuki, W Ludwig et al. (New York, NY: Springer), 1039–1057.
Genilloud, O. (2014). The re-emerging role of microbial natural products in antibiotic discovery. Antonie Van Leeuwenhoek 106, 173–188. doi: 10.1007/s10482-014-0204-6
Glick, B. R. (2015). Beneficial Plant-bacterial Interactions. Cham: Springer. doi: 10.1007/978-3-319-13921-0
Goudjal, Y., Toumatia, O., Sabaou, N., Barakate, M., Mathieu, F., and Zitouni, A. (2013). Endophytic actinomycetes from spontaneous plants of Algerian Sahara: indole-3-acetic acid production and tomato plants growth promoting activity. World J. Microbiol. Biotechnol. 29, 1821–1829. doi: 10.1007/s11274-013-1344-y
Goudjal, Y., Toumatia, O., Yekkour, A., Sabaou, N., Mathieu, F., and Zitouni, A. (2014). Biocontrol of Rhizoctonia solani damping-off and promotion of tomato plant growth by endophytic actinomycetes isolated from native plants of Algerian Sahara. Microbiol. Res. 169, 59–65. doi: 10.1016/j.micres.2013.06.014
Hasegawa, T., Lechevalier, M. P., and Lechevalier, H. A. (1978). A new genus of Actinomycetales: Actinosynnema gen. nov. Int. J. Syst. Bacteriol. 28, 304–310. doi: 10.1099/00207713-28-2-304
Hirsch, A. M., Alvarado, J., Bruce, D., Chertkov, O., De Hoff, P. L., Detter, J. C., et al. (2013). Complete genome sequence of Micromonospora strain L5, a potential plantgrowth regulating actinomycete, originally isolated from Casuarina equisetifolia root nodules. Genome Announc. 1:e00759. doi: 10.1128/genomeA.00759-13
Hoque, M. S., Broadhurst, L. M., and Thrall, P. H. (2011). Genetic characterization of root-nodule bacteria associated with Acacia salicina and A. stenophylla (Mimosaceae) across southeastern Australia. Int. J. Syst. Evol. Microbiol. 61, 299–309. doi: 10.1099/ijs.0.021014-0
Hurek, T., Handley, L. L., Reinhold-Hurek, B., and Piché, Y. (2002). Azoarcus grass endophytes contribute fixed nitrogen to the plant in an unculturable state. Mol. Plant Microbe Interact. 15, 233–242. doi: 10.1094/MPMI.2002.15.3.233
Igarashi, Y., Trujillo, M. E., Martínez-Molina, E., Yanase, S., Miyanaga, S., Obata, T., et al. (2007). Antitumor anthraquinones from an endophytic actinomycete Micromonospora lupini sp. nov. Bioorg. Med. Chem. Lett. 17, 3702–3705. doi: 10.1016/j.bmcl.2007.04.039
Igarashi, Y., Yanase, S., Sugimoto, K., Enomoto, M., Miyanaga, S., Trujillo, M. E., et al. (2011). Lupinacidin C, an inhibitor of tumor cell invasion from Micromonospora lupini. J. Nat. Prod. 74, 862–865. doi: 10.1021/np100779t
Jensen, H. L. (1932). Contributions to our knowledge of the Actinomycetales. III. Further observation on the genus Micromonospora. N.W.S. Proc. Linnean Soc. 57, 173–180.
Jiang, Y., Huang, Y.-H., and Long, Z.-E. (2015). De Novo whole-genome sequence of Micromonospora carbonacea JXNU-1 with broad-spectrum antimicrobial activity, isolated from soil samples. Genome Announc. 3:e00174. doi: 10.1128/genomeA.00174-15
Kaewkla, O., and Franco, C. M. (2013). Rational approaches to improving the isolation of endophytic actinobacteria from Australian native trees. Microb. Ecol. 65, 384–393. doi: 10.1007/s00248-012-0113-z
Kim, T. U., Cho, S. H., Han, J. H., Shin, Y. M., Lee, H. B., and Kim, S. B. (2012). Diversity and physiological properties of root endophytic actinobacteria in native herbaceous plants of Korea. J. Microbiol. 50, 50–57. doi: 10.1007/s12275-012-1417-x
Krause, A., Ramakumar, A., Bartels, D., Battistoni, F., Bekel, T., Boch, J., et al. (2007). Complete genome of the mutualistic, N2-fixing grass endophyte Azoarcus sp. strain BH72. Nat. Biotechnol. 24, 1385–1391. doi: 10.1038/nbt1243
Kunoh, H. (2002). Endophytic actinomycetes:Attractive biocontrol agents. J. Gen. Plant. Pathol. 68, 249–252. doi: 10.1007/PL00013084
Kyeremeth, K., Acquah, K. S., Sazak, A., Houssen, W., Tabudravu, J., Deng, H., et al. (2014). –butremycin, the 3-hydroxyl derivative of ikarugamycin and a protonated aromatic tautomer of 5′-methyltionosine from a Ghanaian Micromonospora sp. K310. Mar. Drugs 12, 999–1012. doi: 10.3390/md12020999
Li, L., Sinkko, H., Montonen, L., Wei, G., Lindström, K., and Räsänen, L. A. (2011). Biogeography of symbiotic and other endophytic bacteria isolated from medicinal Glycyrrhiza species in China. FEMS Microbiol. Ecol. 79, 46–68. doi: 10.1111/j.1574-6941.2011.01198.x
Loria, R., Kers, J., and Joshi, M. (2006). Evolution of plant pathogenicity in Streptomyces. Annu. Rev. Phytopathol. 44, 469–487. doi: 10.1146/annurev.phyto.44.032905.091147
Maldonado, L. A., Fragoso-Yáñez, D., Pérez-García, A., Rosellón-Druker, J., and Quintana, E. T. (2009). Actinobacterial diversity from marine sediments collected in Mexico. Antonie Van Leeuwenhoek 95, 111–120. doi: 10.1007/s10482-008-9294-3
Mark, G. L., Dow, J. M., Kiely, P. D., Higgins, H., Haynes, J., and Baysse, C. (2005). Transcriptome profiling of bacterial responses to root exudates identifies genes involved in microbe-plant interactions. Proc. Natl. Acad. Sci. U.S.A. 102, 17454–17459. doi: 10.1073/pnas.0506407102
Martínez-Hidalgo, P., Galindo-Villardón, P., Igual, J. M., Trujillo, M. E., and Martínez-Molina, E. (2014). Micromonospora from nitrogen fixing nodules of alfalfa (Medicago sativa L.). A new promising Plant Probiotic Bacteria. Sci. Rep. 4:6389. doi: 10.1038/srep06389
Mastronunzio, J. E., Tisa, L. S., Normand, P., and Benson, D. R. (2008). Comparative secretome analysis suggests low plant cell wall degrading capacity in Frankia symbionts. BMC Genomics 9:47 doi: 10.1186/1471-2164-9-47
Mba Medie, M., Davies, G. J., Drancourt, M., and Henrissat, B. (2012). Genome analyses highlight the different biological roles of cellulases. Nat. Rev. Microbiol. 10, 227–234. doi: 10.1038/nrmicro2729
Miao, V., and Davies, J. (2010). Actinobacteria: the good, the bad, and the ugly. Antonie Van Leeuwenhoek 98, 143–150. doi: 10.1007/s10482-010-9440-6
Mingma, R., Phatom-Aree, W., Trakulnaleamsai, S., Thamchaipenet, A., and Duangmal, K. (2014). Isolation of rhizospheric and roots endophytic actinomycetes from Leguminosae plant and their activities to inhibit soybean pathogen, Xanthomonas campestris pv. glycine. World J. Microbiol. Biotechnol. 30, 271–280. doi: 10.1007/s11274-013-1451-9
Misk, A., and Franco, C. C. (2011). Biocontrol of chickpea root rot using endophytic actinobacteria. Biocontrol 56, 811–822. doi: 10.1007/s10526-011-9352-z
Morrissey, J. P., Dow, J. M., Mark, G. L., and O'Gara, F. (2004). Are microbes at the root of a solution to world food production. EMBO Rep. 5, 922–926. doi: 10.1038/sj.embor.7400263
Mukherjee, P. K., Buensanteai, N., Moran-Diez, M. E., Druzhinina, I. S., and Kenerley, C. M. (2012). Functional analysis of non-ribosomal peptide synthetases (NRPSs) in Trichoderma virens reveals a polyketide synthase (PKS)/NRPS hybrid enzyme involved in the induced systemic resistance response in maize. Microbiology 158, 155–165. doi: 10.1099/mic.0.052159-0
Müller, H., Berg, C., Landa, B. B., Auerbach, A., Moissl-Eichinger, C., and Berg, C. (2015). Plant genotype-specific archaeal and bacterial endophytes but similar Bacillus antagonists colonize Mediterranean olive trees. Front. Microbiol. 6:138. doi: 10.3389/fmicb.2015.00138
Muresu, R., Polone, E., Sulas, L., Baldan, B., Tondello, A., Delogu, G., et al. (2008). Coexistence of predominantly nonculturable rhizobia with diverse, endophytic bacterial taxa within nodules of wild legumes. FEMS Microbiol. Ecol. 63, 383–400. doi: 10.1111/j.1574-6941.2007.00424.x
Okubo, T., Ikeda, S., Sasaki, K., Ohshima, K., Hattori, M., Sato, T., et al. (2014). Phylogeny and functions of bacterial communities associated with field-grown rice shoots. Microbes Environ. 29, 329–332. doi: 10.1264/jsme2.ME14077
Ørskov, J. (1923). Investigations into the Morphology of Ray Fungi. Copenhagen: Levin and Munskgaard.
Pinto, C., Pinho, D., Sousa, S., Pinheiro, M., Egas, C., and Gomes, A. C. (2014). Unravelling the diversity of grapevine microbiome. PLoS ONE 9:e85622. doi: 10.1371/journal.pone.0085622
Pujic, P., Fournier, P., Alloisio, N., Hay, A. E., Maréchal, J., Anchisi, S., et al. (2012). Lectin genes in the Frankia alni genome. Arch. Microbiol. 194, 47–56. doi: 10.1007/s00203-011-0770-1
Qin, S., Chen, H. H., Zhao, G. Z., Li, J., Zhu, W. Y., Xu, L. H., et al. (2012). Abundant and diverse endophytic actinobacteria associated with medicinal plant Maytenus austroyunnanensis in Xishuangbanna tropical rainforest revealed by culture-dependent and culture-independent methods. Environ. Microbiol. Rep. 4, 522–531. doi: 10.1111/j.1758-2229.2012.00357.x
Qin, S., Li, J., Chen, H. H., Zhao, G. Z., Zhu, W. Y., Jiang, C. L., et al. (2009). Isolation, diversity, and antimicrobial activity of rare actinobacteria from medicinal plants of tropical rain forests in Xishuangbanna, China. Appl. Environ. Microbiol. 75, 6176–6186. doi: 10.1128/AEM.01034-09
Qin, S., Xing, K., Jiang, J.-H., Xu, L.-H., and Li, W.-J. (2011). Biodiversity, bioactive natural products and biotechnological potential of plant-associated endophytic actinobacteria. Appl. Microbiol. Biotechnol. 89, 457–473. doi: 10.1007/s00253-010-2923-6
Reid, A., and Greene, S. E. (2012). How Microbes Help Feed the World. Washington, DC: American Academy of Microbiology.
Rhoden, S. A., Garcia, A., Santos e Silva, M. C., Azevedo, J. L., and Pamphile, J. A. (2015). Phylogenetic analysis of endophytic bacterial isolates from leaves of the medicinal plant Trichilia elegans A. Juss. (Meliaceae). Genet. Mol. Res. 14, 1515–1525. doi: 10.4238/2015.February.20.7
Robledo, M., Jiménez-Zurdo, J. I., Velázquez, E., Trujillo, M. E., Zurdo-Piñeiro, J. L., Ramírez-Bahena, M. H., et al. (2008). Rhizobium cellulase CelC2 is essential for primary symbiotic infection of legume host roots. Proc. Natl. Acad. Sci. U.S.A. 105, 7064–7069. doi: 10.1073/pnas.0802547105
Robledo, M., Rivera, L., Jiménez-Zurdo, J. I., Rivas, R., Dazzo, F., Velázquez, E., et al. (2012). Role of Rhizobium endoglucanase CelC2 in cellulose biosynthesis and biofilm formation on plant roots and abiotic surfaces. Microb. Cell Fact. 11:125. doi: 10.1186/1475-2859-11-125
Rodríguez, R. (2008). Análisis de la Población Bacteriana Presente en Nódulos de Lupinus: Interacción y Localización in Situ. Ph.D. thesis, Universidad de Salamanca, Salamanca.
Rungin, S., Indananda, C., Suttiviriya, P., Kruasuwan, W., Jamsaeng, R., and Thamchaipenet, A. (2012). Plant growth enhancing effects by a siderophore-producing endophytic streptomycete isolated from a Thai jasmine rice plant (Oryza sativa L. cv, KDML105). Antonie Van Lwwuwenhoek 102, 463–472. doi: 10.1007/s10482-012-9778-z
Ryan, R. P., Germaine, K., Franks, A., and Ryan, D. J. (2008). Bacterial endophytes: recent developments and applications. FEMS Microbiol. 278, 1–9. doi: 10.1111/j.1574-6968.2007.00918.x
Saddler, G. S. Messenber-Guimaraes (2012). “Genus Curtobacterium,” in Bergey's Manual of Systematic Bacteriology, Vol. 5, eds M. Goodfellow, P. Kämpfer, H.-J. Busse, M. E. Trujillo, K.-I. Suzuki, W Ludwig, et al. (New York, NY: Springer), 887–895.
Sardi, P., Saracchi, M., Quaroni, S., Petrolini, B., Borgonovi, G. E., and Merli, S. (1992). Isolation of endophytic Streptomyces strains from surface-sterilized roots. Appl. Environ. Microbiol. 58, 2691–2693.
Schenk, P. M., Carvalhais, L. C., and Kazan, K. (2012). Unraveling plant–microbe interactions: can multi-species transcriptomics help? Trends Biotecnol. 30, 177–184. doi: 10.1016/j.tibtech.2011.11.002
Shidore, T., Dinse, T., Öhrlein, J., Becker, A., and Reinhold-Hurek, B. (2012). Transcriptomic analysis of responses to exudates reveal genes required for rhizosphere competence of the endophyte Azoarcus sp. strain BH72. Environ. Microbiol. 14, 2775–2787. doi: 10.1111/j.1462-2920.2012.02777.x
Solans, M. (2007). Discaria trinervis—Frankia symbiosis promotion by saprohytic actinomycetes. J. Basic Microbiol. 47, 243–250. doi: 10.1002/jobm.200610244
Solans, M., Vobis, G., Cassán, F., Luna, V., and Wall, L. G. (2011). Production of phytohormones by root-associated saprophytic actinomycetes isolated from the actinorhizal plant Ochetophila trinervis. World J. Microbiol. Biotechnol. 27, 2195–2202. doi: 10.1007/s11274-011-0685-7
Solans, M., Vobis, G., and Wall, L. G. (2009). Actinomycetes promote nodulation in Medicago sativa-Sinorhizobium meliloti simbiosis in the presence of high N. J. Plant Growth Regul. 28, 106–114. doi: 10.1007/s00344-009-9080-0
Stajković, O., de Meyer, S., Mili, B., Willems, A., and Deli, D. (2009). Isolation and characterization of endophytic non-rhizobial bacteria from root nodules of alfalfa (Medicago sativa L.). Bot. Serb. 33, 107–114.
Sturz, A. V., Christie, B. R., Matheson, B. G., and Nowak, J. (1997). Biodiversity of endophytic bacteria which colonize red clover nodules, roots, stems and foliage and their influence on host growth. Biol. Fertil. Soils. 25, 13–19. doi: 10.1007/s003740050273
Taghavi, S., van der Lelie, D., Hoffman, A., Zhang, Y. B., Walla, M. D., Vangronsveld, J., et al. (2010). Genome sequence of the plant growth promoting endophytic bacterium Enterobacter sp. 638. PLoS Genet. 6:e1000943. doi: 10.1371/journal.pgen.1000943
Thawai, C., Kittakoop, P., Tanasupawat, S., Suwanborirux, K., Sriklun, K., and Thebtaranonth, Y. (2004). Micromonosporin A, a novel 24-membere polyene lactam macrolide from Micromonospora sp. isolated from peat swamp forest. Chem. Biodivers. 1, 640–645. doi: 10.1002/cbdv.200490055
Tokala, R. K., Strap, J. L., Jung, C. M., Crawford, D. L., Salove, M. H., Deobald, L. A., et al. (2002). Novel plant-microbe rhizosphere interaction involving Streptomyces lydicus WYEC108 and the pea plant (Pisum sativum). Appl. Environ. Microbiol. 68, 2161–2171. doi: 10.1128/AEM.68.5.2161-2171.2002
Trujillo, M. E., Alonso-Vega, P., Rodríguez, R., Carro, L., Cerda, E., Alonso, P., et al. (2010). The genus Micromonospora is widespread in legume root nodules: the example of Lupinus angustifolius. ISME J. 4, 1265–1281. doi: 10.1038/ismej.2010.55
Trujillo, M. E., Bacigalupe, R., Pujic, P., Igarashi, Y., Benito, P., Riesco, R., et al. (2014b). Genome features of the endophytic actinobacterium Micromonospora lupini strain Lupac 08: on the process of adaptation to an endophytic life style? PLoS ONE 9:e108522. doi: 10.1371/journal.pone.0108522
Trujillo, M. E., Hong, K., and Genilloud, O. (2014a). “Family Micromonosporaceae,” in The Prokaryotes, The Actinobacteria, eds E. Ronsenberg, E. F. DeLong, S. Lory, E. Stackebrandt, and F. Thompson (Heidelberg: Springer), 499–569. doi: 10.1007/978-3-642-30138-4_196
Trujillo, M. E., Kroppenstedt, R. M., Fernández-Molinero, C., Schumann, P., and Martínez-Molina, E. (2007). Micromonospora lupini sp. nov. and Micromonospora saelicesensis sp. nov., isolated from root nodules of Lupinus angustifolius. Int. J. Syst. Evol. Microbiol. 57, 2799–2804. doi: 10.1099/ijs.0.65192-0
Trujillo, M. E., Kroppenstedt, R. M., Schumann, P., Carro, L., and Martinez-Molina, E. (2006). Micromonospora coriariae sp. nov., isolated from root nodules of Coriaria myrtifolia. Int. J. Syst. Evol. Microbiol. 56, 2381–2385. doi: 10.1099/ijs.0.64449-0
Udwary, D. W., Gontang, E. A., Jones, A. C., Jones, C. S., Schultz, A. W., Winter, J. M., et al. (2011). Significant natural product biosynthetic potential of actinorhizal symbionts of the genus Frankia, as revealed by comparative genomic and proteomic analyses. Appl. Environ. Microbiol. 77, 3617–3625. doi: 10.1128/AEM.00038-11
Valdés, M., Perez, N. O., Estrada de Los Santos, P., Caballero-Mellado, J., Peña-Cabriales, J. J., Normand, P., et al. (2005). Non-Frankia actinomycetes isolated from surfacesterilized roots of Casuarina equisetifolia fix nitrogen. Appl. Environ. Microbiol. 71, 460–466. doi: 10.1128/AEM.71.1.460-466.2005
Verma, V. C., Gond, S. K., Kumar, A., Mishra, A., Kharwar, R. N., and Gange, A. C. (2009). Endophytic actinomycetes from Azadirachta indica A. Juss.: isolation, diversity, and anti-microbial activity. Microbiol. Ecol. 57, 749–756. doi: 10.1007/s00248-008-9450-3
Verma, V. C., Singh, S. K., and Prakash, S. (2011). Bio-control and plant growth promotion potential of siderophore producing endophytic Streptomyces from Azadirachta indica A. Juss. J. Basic Microbiol. 51, 550–556. doi: 10.1002/jobm.201000155
Vincent, J. M. (1970). “The cultivation, isolation and maintenance of rhizobia,” in A Manual for the Practical Study of Root Nodule Bacteria, ed J. M. Vincent (Oxford: Blackwell Scientific), 1–13.
Weinstein, M. J., Luedemann, G. M., Oden, E. M., Wagman, G. H., Rosselet, J. P., Marquez, J. A., et al. (1963). Gentamicin, a new antibiotic complex from Micromonospora. Med. Chem. 6, 463–464. doi: 10.1021/jm00340a034
Yano, K., Wada, T., Suzuki, S., Tagami, K., Matsumoto, T., Shiwa, Y., et al. (2013). Multiple rRNA operons are essential for efficient cell growth and sporulation as well as outgrowth in Bacillus subtilis. Microbiology 159, 2225–2236. doi: 10.1099/mic.0.067025-0
Young, A. J., Petrasovits, L. A., Croft, B. J., Gillings, M., and Brumbley, S. M. (2006). Genetic uniformity of international isolates of Leifsonia xyli subsp. xyli, causal agent of ratoon stunting disease of sugarcane. Austr. Plant Pathol. 35, 503–511. doi: 10.1071/AP06055
Zakhia, F., Jeder, H., Willems, A., Gillis, M., Dreyfus, B., and de Lajudie, P. (2006). Diverse bacteria associated with root nodules of spontaneous legumes in Tunisia and first report for nifH-like gene within the genera Microbacterium and Starkeya. Microb. Ecol. 51, 375–393. doi: 10.1007/s00248-006-9025-0
Keywords: Micromonospora, legumes, PGPB, actinorhizal, endophytic, nodule
Citation: Trujillo ME, Riesco R, Benito P and Carro L (2015) Endophytic Actinobacteria and the Interaction of Micromonospora and Nitrogen Fixing Plants. Front. Microbiol. 6:1341. doi: 10.3389/fmicb.2015.01341
Received: 21 July 2015; Accepted: 16 November 2015;
Published: 01 December 2015.
Edited by:
Sheng Qin, Jiangsu Normal University, ChinaReviewed by:
James A. Coker, University of Maryland University College, USAJulia Maresca, University of Delaware, USA
Copyright © 2015 Trujillo, Riesco, Benito and Carro. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Martha E. Trujillo, mett@usal.es
 Martha E. Trujillo
Martha E. Trujillo Raúl Riesco
Raúl Riesco Patricia Benito
Patricia Benito Lorena Carro
Lorena Carro