- 1Department of Plant Biotechnology and Bioinformatics, Ghent University, Ghent, Belgium
- 2VIB Center for Plant Systems Biology, Ghent, Belgium
Wood is a renewable resource that is mainly composed of lignin and cell wall polysaccharides. The polysaccharide fraction is valuable as it can be converted into pulp and paper, or into fermentable sugars. On the other hand, the lignin fraction is increasingly being considered a valuable source of aromatic building blocks for the chemical industry. The presence of lignin in wood is one of the major recalcitrance factors in woody biomass processing, necessitating the need for harsh chemical treatments to degrade and extract it prior to the valorization of the cell wall polysaccharides, cellulose and hemicellulose. Over the past years, large research efforts have been devoted to engineering lignin amount and composition to reduce biomass recalcitrance toward chemical processing. We review the efforts made in forest trees, and compare results from greenhouse and field trials. Furthermore, we address the value and potential of CRISPR-based gene editing in lignin engineering and its integration in tree breeding programs.
Introduction
Fossil resources are the main feedstock for energy and organic compounds, and their use results in the emission of greenhouse gases associated with climate change. The coming climate crash calls for an urgent transition from a fossil-based to a bio-based economy in which lignocellulosic biomass rather than oil is used for the production of fuels, chemicals and materials. Wood is an important source of lignocellulosic biomass; it is mainly composed of secondary-thickened cell walls rich in cellulose, hemicelluloses, and lignin. All three polymers can be valorized in the bio-based economy. Cellulose is a source for the pulp and paper industry, and both cellulose and hemicelluloses can be depolymerized to their monosaccharides for fermentation into, e.g., bio-ethanol, lactic acid and detergents (Vanholme et al., 2013b). As lignin negatively affects the efficiency of wood processing toward these applications, trees can be engineered to accumulate less lignin, to become more amenable for the production of paper and fermentable sugars. On the other hand, lignin is increasingly being considered a valuable component in the bio-based economy. Indeed, given that lignin is the largest renewable aromatic source on Earth, the economic viability of a bio-refinery can be significantly increased if lignin is also valorized, and used as a resource for the production of chemicals (Holladay et al., 2007; Tuck et al., 2012; Davis et al., 2013; Ragauskas et al., 2014; Li C. et al., 2015; Van den Bosch et al., 2015; Rinaldi et al., 2016; Upton and Kasko, 2016; Schutyser et al., 2018).
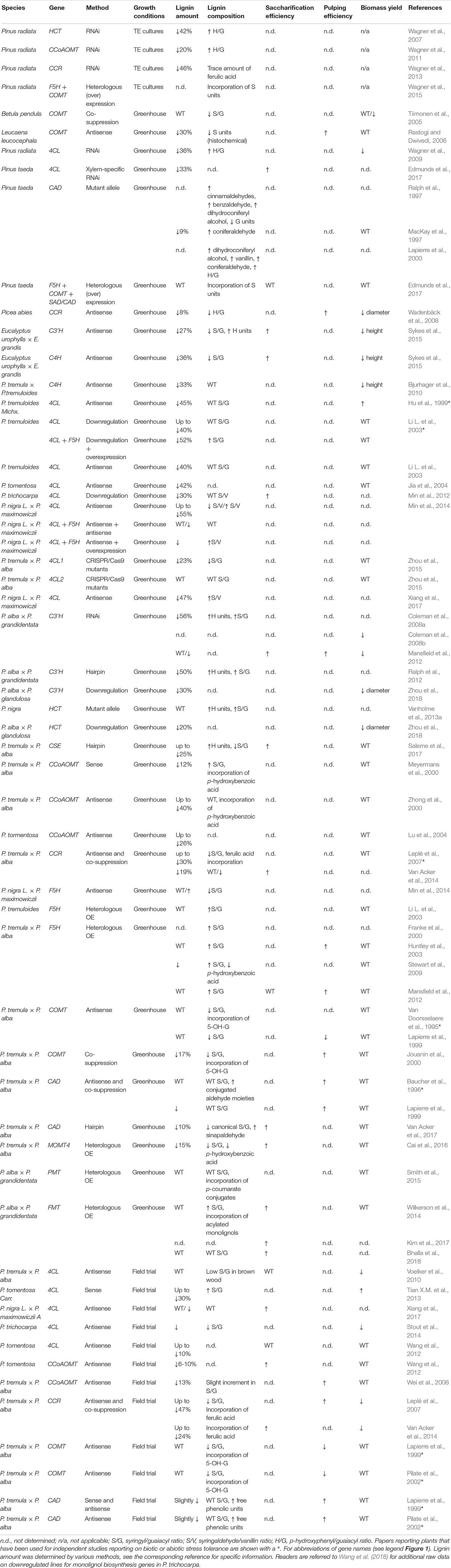
The lignin polymer is composed of monolignols that are produced by the phenylpropanoid and monolignol biosynthetic pathways, by a series of enzymatic reactions starting with the deamination of phenylalanine (Figure 1). The monolignols are synthesized in the cytoplasm and translocated to the apoplast, where they are dehydrogenated to monolignol radicals by the action of laccases and peroxidases (Berthet et al., 2011; Zhao et al., 2013). These monolignol radicals then couple with each other in a combinatorial fashion, generating a range of chemical bonds such as the aryl-ether bond (β-O-4), resinol bond (β-β), and phenylcoumaran bond (β-5) (Boerjan et al., 2003; Ralph et al., 2004; Vanholme et al., 2010). The most common monolignols are the hydroxycinnamyl alcohols p-coumaryl, coniferyl, and sinapyl alcohols, which generate the H, G, and S units upon their incorporation into the lignin polymer, respectively (Bonawitz and Chapple, 2010; Ralph et al., 2019; Vanholme et al., 2019). The relative contribution of the lignin building blocks varies among taxa, developmental stage, tissue and cell type, and even cell wall layer; lignin from softwoods (gymnosperms) is comprised almost entirely of G units with a minor fraction of H units, while lignin from hardwoods (angiosperms) has S units in addition to G units and traces of H units (Boerjan et al., 2003; Vanholme et al., 2010, 2019). Besides these traditional monolignols, a variety of other p-hydroxylated aromatic molecules can be incorporated in the lignin polymer to various levels (Vanholme et al., 2019).
Figure 1. Lignin biosynthetic pathway. Alternative monomers and heterologously expressed enzymes are shown in bold. PAL, phenylalanine ammonia-lyase; C4H, cinnamate 4-hydroxylase; 4CL, 4-coumarate:CoA ligase; HCT, p-Hydroxycinnamoyl-CoA:quinate/shikimate-p-hydroxycinnamoyltransferase; C3’H, p-coumaroyl quinate/shikimate 3’-hydroxylase; CSE, caffeoyl shikimate esterase; CCoAOMT, caffeoyl-CoA O-methyltransferase; CCR, cinnamoyl-CoA reductase; F5H, ferulate 5-hydroxylase/CAld5H, coniferaldehyde 5-hydroxylase; COMT, caffeic acid O-methyltransferase; CAD, cinnamyl alcohol dehydrogenase; FMT, feruloyl-CoA monolignol transferase; PMT, p-coumaroyl-CoA monolignol transferase.
Given that lignin is a major recalcitrance factor in wood delignification processes, large research efforts have been devoted to unravel the lignin biosynthetic pathway, and to study the effects of perturbations of the lignin biosynthesis genes on lignin amount and composition, and on wood processing efficiency. While modifications in genes ranging from those encoding transcription factors up to those encoding oxidative enzymes have resulted in altered lignin content, composition or deposition (Eriksson et al., 2000; Li Y.H. et al., 2003; Liang et al., 2008; Lu et al., 2013; Lin et al., 2016; Xu et al., 2017; Yang et al., 2017; Obudulu et al., 2018), this review will focus on the results obtained by engineering the lignin biosynthetic genes.
Engineering the Lignin Pathway
Table 1 provides an overview of the different studies on downregulated or mutated lignin biosynthetic genes in poplar, pine, eucalyptus and birch, with the resulting effects on wood processing efficiencies, when determined. Reducing the activity of any step of the lignin biosynthetic pathway, starting from PAL up to CAD may result in a reduction in lignin content (Table 1). Several parameters influence the degree of lignin reduction, such as the target gene and the degree of downregulation of the enzyme activity, which in turn depends on the efficiency of the silencing construct used, the size of the gene family, and redundancy within the gene family. Generally, the downregulation of the steps from C4H up to CCR results in a more dramatic reduction in lignin amount (Hu et al., 1999; Meyermans et al., 2000; Zhong et al., 2000; Li L. et al., 2003; Jia et al., 2004; Lu et al., 2004; Leplé et al., 2007; Coleman et al., 2008a,b; Bjurhager et al., 2010; Mansfield et al., 2012; Ralph et al., 2012; Min et al., 2014; Van Acker et al., 2014; Zhou et al., 2015, 2018; Saleme et al., 2017; Xiang et al., 2017) than downregulation of F5H, COMT and CAD (Van Doorsselaere et al., 1995; Baucher et al., 1996; Lapierre et al., 1999; Jouanin et al., 2000; Van Acker et al., 2014; Wang et al., 2018). Lignin reduction can be associated with an increase in S/G, such as in C3’H- (Coleman et al., 2008a; Ralph et al., 2012) and CCoAOMT-downregulated trees (Meyermans et al., 2000), or a decrease in S/G ratio such as in CSE- (Saleme et al., 2017), and COMT-downregulated trees (Van Doorsselaere et al., 1995; Lapierre et al., 1999; Jouanin et al., 2000). Interestingly, low-lignin 4CL-downregulated poplars were found to have an increase in S/G (Min et al., 2014; Xiang et al., 2017), a decrease in S/G (Voelker et al., 2010; Zhou et al., 2015), or ratios comparable to wild type (Hu et al., 1999; Li L. et al., 2003). This variance cannot be associated with the promoter or the method used for downregulation, suggesting that differences in the degree of silencing, growth conditions or developmental state influence this trait. On the other hand, the strongest effects on H/G/S lignin composition have been observed for trees downregulated in C3’H and HCT, which deposit lignin with large increases in H unit content (Coleman et al., 2008a; Ralph et al., 2012; Vanholme et al., 2013a), whereas trees that overexpress F5H produce lignin strongly enriched in S units (Franke et al., 2000; Li L. et al., 2003; Stewart et al., 2009), and trees that are downregulated in COMT have dramatically reduced S unit biosynthesis (Van Doorsselaere et al., 1995; Lapierre et al., 1999).
Both the reduced lignin content and variation in the H/G/S ratios can affect the biomass processing efficiency. Consistent with the established role of lignin in determining biomass recalcitrance (Zeng et al., 2014; McCann and Carpita, 2015; Li et al., 2016; Wang et al., 2018), plants with reduced levels of lignin show increased chemical pulping and saccharification efficiency (Hu et al., 1999; Jouanin et al., 2000; Rastogi and Dwivedi, 2006; Wadenbäck et al., 2008; Wang et al., 2012; Sykes et al., 2015; Cai et al., 2016; Edmunds et al., 2017; Saleme et al., 2017; Xiang et al., 2017; Van Acker et al., 2017; Wang et al., 2018). An increased level of H units reduces lignin polymer length and, hence, increases the removal of lignin from the biomass (Mansfield et al., 2012; Sykes et al., 2015). Increased S/G results in lignin more easily cleaved and extracted in alkaline conditions, supposedly due to the lower degree of polymerization (Huntley et al., 2003; Stewart et al., 2009; Mansfield et al., 2012; Yoo et al., 2018).
The processing efficiency of the biomass can also be modified by the increased incorporation of molecules that generally represent minor components in the lignin of wild-type plants. The incorporation of ferulic acid in CCR-deficient trees results in the formation of acetal bonds in the lignin polymer, which are easily cleaved in acidic biomass pretreatments (Leplé et al., 2007; Ralph et al., 2008; Van Acker et al., 2014). Indeed, the levels of ferulic acid in lignin positively correlated with a higher saccharification efficiency (Van Acker et al., 2014). The incorporation of 5-hydroxyconiferyl alcohol and 5-hydroxyconiferaldehyde in the lignin of COMT-deficient poplars (Van Doorsselaere et al., 1995; Lapierre et al., 1999; Jouanin et al., 2000; Morreel et al., 2004; Lu et al., 2010) gives rise to benzodioxane bonds, potentially preventing covalent linkages between lignin and the polysaccharide hydroxyl groups (Weng et al., 2010; Vanholme et al., 2012; Nishimura et al., 2018). On the other hand, COMT deficiency also results in a more condensed lignin due to the relatively higher levels of the condensed β-β and β-5 bonds, and the lower levels of β-O-4 bonds, when the S unit frequency drops. Chemical pulping of wood derived from poplars strongly downregulated for COMT resulted in a higher pulp yield, counterbalanced by the residual lignin content in the pulp. These trees had a lower lignin and a higher cellulose content (Jouanin et al., 2000). On the other hand, poplars that were modestly downregulated for COMT had a large decrease in pulp yield, presumably because lignin content had remained normal while the lignin had a higher frequency of condensed bonds that negatively affected the lignin extraction (Lapierre et al., 1999; Pilate et al., 2002). The incorporation of cinnamaldehydes in the lignin polymer in CAD-deficient trees results in shorter lignin polymer chains, hence a higher proportion of free phenolic end groups that increase the solubility of the polymer in alkali. The incorporation of cinnamaldehydes in the lignin polymer presumably also reduces the covalent interaction of the aliphatic chain with hemicellulose, again rendering the lignin more soluble. In addition, due to the extended conjugated system that is generated when a cinnamaldehyde β-O-4 couples with another monomer, the aromatic ether bond of the incorporated cinnamaldehyde becomes more susceptible to alkaline cleavage (Lapierre et al., 1989; Van Acker et al., 2017).
Lignin polymerization is a combinatorial radical coupling process, allowing a wide range of phenolic compounds to be naturally incorporated into the lignin polymer (Boerjan et al., 2003; Vanholme et al., 2019). Researchers have attempted to tailor the lignin amount and composition to improve biomass processing by expression of heterologous genes, aiming at the biosynthesis and incorporation of various compatible phenolic compounds as alternative monolignols into the lignin polymer (Ralph, 2006; Vanholme et al., 2012; Mottiar et al., 2016; Mahon and Mansfield, 2018). One example is the introduction of genes encoding enzymes that are needed for S unit biosynthesis in pine; the simultaneous expression of F5H, COMT and CAD successfully introduced S units in Pinus radiata (Wagner et al., 2015; Edmunds et al., 2017). The introduction of the gene encoding a monolignol 4-O-methyltransferase (MOMT4) into poplar leads to the formation of 4-O-methylated coniferyl and sinapyl alcohols, which cannot be incorporated into the growing lignin polymer because they lack the aromatic hydroxyl group. This leads to a halt in lignin polymerization and results in trees with lower lignin content and higher saccharification efficiency (Bhuiya and Liu, 2010; Cai et al., 2016). Poplars have also been engineered to contain ester linkages in the lignin polymer backbone. Coniferyl ferulate esters were introduced into the polymer via expression of a FERULOYL-CoA:MONOLIGNOL TRANSFERASE (FMT) gene derived from Angelica sinensis (Wilkerson et al., 2014), leading to an improved saccharification efficiency under various pretreatment conditions (Wilkerson et al., 2014; Kim et al., 2017; Bhalla et al., 2018), and an improved kraft pulping efficiency as compared to wild type (Zhou et al., 2017). Monolignol p-coumarate esters have also been engineered in poplar, via expression of a rice p-COUMAROYL-CoA:MONOLIGNOL TRANSFERASE (PMT) gene, resulting in a higher frequency of resistant interunit bonds and a higher frequency of G and S terminal units with free phenolic groups (Smith et al., 2015; Sibout et al., 2016). While in Arabidopsis the heterologous expression of PMT resulted in a reduced lignin amount accompanied by an increased saccharification efficiency (Sibout et al., 2016), there was no decrease in lignin amount in poplar and the saccharification efficiency was not determined (Smith et al., 2015).
While several modifications of the lignin amount and composition were shown to provide improvements in biomass processing, these modifications were often accompanied by a biomass yield penalty (Leplé et al., 2007; Wadenbäck et al., 2008; Wagner et al., 2009; Voelker et al., 2010; Stout et al., 2014; Van Acker et al., 2014; Sykes et al., 2015; Zhou et al., 2018). A recent metastudy perturbed 21 lignin biosynthesis genes in P. trichocarpa, and comprehensively integrated the results of transcriptomic, proteomic, fluxomic, and phenomic data of 221 lines. The authors concluded that tree growth is not associated with lignin amount, subunit composition or specific linkages (Wang et al., 2018), but rather correlated with the presence of collapsed xylem vessels (Coleman et al., 2008a,b; Wagner et al., 2009; Voelker et al., 2010; Vargas et al., 2016; De Meester et al., 2018), the activation of a cell wall integrity pathway (Bonawitz et al., 2014) and/or the accumulation of chemical inhibitors (Gallego-Giraldo et al., 2011; Muro-Villanueva et al., 2019).
Whereas substantial efforts have been made to decrease lignin content by downregulation of lignin biosynthetic genes, studies on the upregulation of the lignin pathway and the overproduction of lignin have been scarce. Indeed, reports on the overexpression of F5H show an unchanged or even a decrease in lignin content (Huntley et al., 2003; Li L. et al., 2003; Stewart et al., 2009; Mansfield et al., 2012; Edmunds et al., 2017). The overexpression of CAD and COMT has resulted in gene-silencing rather than upregulation, or no effect on expression levels was detected (Baucher et al., 1996; Lapierre et al., 1999; Jouanin et al., 2000; Leplé et al., 2007; Van Acker et al., 2014). The overexpression of the R2R3-MYB transcription factors PtoMYB92, PtoMYB216, and PtoMYB74 all resulted in additional xylem layers, thicker xylem cell walls as well as ectopic lignin deposition, and the plants accumulated 13–50% more lignin (Tian Q. et al., 2013; Li C.F. et al., 2015; Li et al., 2018). The MYB overexpression lines constitutively upregulated the lignin biosynthesis pathway genes, and while plants overexpressing MYB92 and MYB74 had a biomass penalty, the overexpression of MYB216 resulted in plants with up to 50% more lignin and no developmental phenotype. As lignin is increasingly being considered an important resource for the sustainable production of chemicals (Cao et al., 2018) the engineering of plants overproducing lignin should be further explored.
Field Trials
The examples discussed above clearly show that lignin engineering via down- or upregulation of phenylpropanoid pathway genes – or expression of heterologous genes – has the potential to increase the processing efficiency of lignocellulosic biomass. Due to practical and regulatory reasons, most studies report on data obtained from the analysis of trees grown in a greenhouse. However, experiments with trees grown in a greenhouse typically do not take into account developmental processes such as growth cessation and dormancy. In addition, greenhouse experiments do not provide sufficient insight into the interaction of the engineered plant with environmental factors such as soil type, wind, and pathogens. Understanding these interactions is an important step in the translation of research results toward commercial applications. Indeed, the body of work produced by studies for which permission to establish field trials was granted, highlights important differences in phenotype between greenhouse- and field grown trees. Table 1 summarizes the reports on field trials performed with 4CL, CCoAOMT, CCR, COMT, and CAD downregulated trees.
Confirming the potential of modified lignocellulosic biomass as a substrate for applications, several lignin-engineered field-trial grown trees showed improvements in wood processing. Poplars downregulated for CCoAOMT grown for 5 years in a field trial in Beijing (China), showed an increased glucose and xylose release upon saccharification (Wang et al., 2012). Poplars downregulated for CCR and grown in a field trial in France, proved to be more amenable to chemical kraft pulping (Leplé et al., 2007). Two additional field trials conducted in France and Belgium with CCR-downregulated poplars resulted in up to 160% improvement in ethanol production in simultaneous saccharification and fermentation (SSF) assays; however the plants had up to 50% biomass reduction (Van Acker et al., 2014). Field trials with CAD-downregulated poplar also showed promising results. These trees showed slightly less lignin than wild type and proved more amenable to kraft delignification (Lapierre et al., 1999). Consistently, the same lines grown in larger-scale field trials in France and the United Kingdom showed a mild decrease in lignin amount and an improvement in kraft pulping deemed commercially relevant, since the plants needed 6% less alkali to achieve a delignification similar to that of wild-type trees (Pilate et al., 2002).
However, conflicting reports on both biomass yield and downstream processing efficiency suggest that these parameters are highly influenced by environmental factors. While a field trial conducted in China using 4CL downregulated poplars found that, even with a 28% decrease in lignin content compared to wild type, the trees had about 8% increased height (Tian X.M. et al., 2013), consistent with greenhouse studies (Hu et al., 1999), other field trials found that 4CL-downregulated poplars had decreased biomass and were sometimes even dwarfed (Voelker et al., 2010; Stout et al., 2014; Marchin et al., 2017). Reports also diverge regarding downstream processing efficiencies of wood derived from these 4CL-downregulated field-grown poplars. While up to 100% increase in sugar recovery was found for 4CL1-downregulated trees (35S-driven antisense 4CL construct) grown in a mountain site in the United States (Xiang et al., 2017), data obtained from field studies conducted in Oregon (United States) found that Pt4CL1 promoter-driven antisense silenced 4CL1 poplars had no improvement in saccharification efficiency compared to wild type (Voelker et al., 2010). Likewise, a long term study in Wenling (China), found that 4CL-downregulated poplars did not show a significant improvement in sugar yield compared to wild type (Wang et al., 2012). In both latter cases, the trees showed mild decreases in lignin amount which did not translate into higher processing efficiency, potentially because of the higher concentration of extractives that could interfere with enzymatic activity (Voelker et al., 2010).
Field trial studies have shown that environmental factors can influence lignification and restore traits to wild-type levels as compared to the levels achieved when the same plants were grown in the greenhouse. While 4CL-downregulated trees had decreased lignin content when grown under greenhouse conditions, analysis of the same 4CL antisense poplars, but grown in the field, has often shown that the lignin content was increased as compared to the greenhouse-grown trees and sometimes even restored to wild-type levels (Stout et al., 2014; Xiang et al., 2017). Similarly, lignin levels were much less reduced in CCR-deficient poplars when they were grown in the field as compared to when they were grown in the greenhouse (Van Acker et al., 2014). At least for the CCR-deficient poplars, it is possible that the higher lignification level of field-grown trees is due to the fact that the wood samples were taken during winter. When tree growth ceases in autumn, the trees still have time to fully lignify their cell walls by the time the tree enters dormancy, as compared to greenhouse-grown trees that develop new xylem continuously. Lignin composition has also been shown to differ between greenhouse- and field-grown low-lignin trees. 4CL-downregulated poplars grown in a field in North Carolina had lignin with a lower S/G than when the same lines were grown in the greenhouse (Stout et al., 2014).
Taken together, these results show that data obtained from greenhouse-grown trees cannot easily be extrapolated to field-grown trees, underpinning the need for field trial experiments at different locations. Some lines presented a yield penalty rendering them less interesting for applications, highlighting the need for a better understanding of the molecular basis of the yield penalty and the development of strategies to overcome this problem.
Lignin has been shown to play an important role in pathogen resistance (Miedes et al., 2014; Zhao and Dixon, 2014), and it plays a pivotal role in allowing the plant to transport water. This suggests that lignin modifications could have an impact on plant stress tolerance. While further investigation is needed to fully address this possibility, the downregulation of 4CL, COMT, and CAD in poplar did not dramatically alter the feeding performance of leaf-feeding herbivores (Tiimonen et al., 2005; Brodeur-Campbell et al., 2006; Hjalten et al., 2013). The effect of the downregulation of COMT and CAD in poplar on plant-insect interactions has also been assessed on field-grown trees, and it was shown that the lignin-modified trees had normal incidence of visiting and feeding insects, as well as normal responses to microbial pathogens (Pilate et al., 2002; Halpin et al., 2007). These results indicate that trees with modified lignin do not necessarily suffer more than wild-type plants from pests and diseases. Nevertheless, profiling of the endosphere bacterial microbiome of wood harvested from field-grown, CCR-downregulated poplars demonstrated shifts in the bacterial community, presumably because of the altered abundance of particular phenolic metabolites in the xylem (Beckers et al., 2017).
Considering the role of lignin in xylem function and structure, the water relations of a few low lignin-modified poplars have been assessed. 4CL-downregulated poplars were found to have reduced hydraulic conductivity, potentially interfering with plant growth (Marchin et al., 2017). Hydraulic stress experiments with poplars downregulated for CCR, COMT or CAD showed that these plants had a lower resistance to cavitation, while maintaining normal xylem hydraulic conductivity and water transport (Awad et al., 2012). These results suggest that the growth of low-lignin mutants might be influenced by water availability. As for any new hybrid obtained from classical breeding, field tests are needed to evaluate field performance and stress tolerance of lignin-engineered trees.
Prospects for Lignin Engineering in Forest Trees
The performance of lignin-engineered plants appears to be highly influenced by the environmental conditions. It is unclear, however, whether the differences observed between greenhouse-grown and field-grown trees, or between trees grown in different field locations, result from different levels of gene suppression or from interaction of the engineered trait with the environment (GxE). Indeed, unstable downregulation is a shortcoming of gene silencing techniques that are based on RNAi. This is witnessed by observing variation in the red xylem phenotype that is observed when particular lignin biosynthesis genes, such as CAD, COMT, or CCR, are downregulated. The red xylem coloration is often not uniform throughout the xylem, but rather appears in patches that reflect variable levels of gene silencing (Leplé et al., 2007; Voelker et al., 2010; Van Acker et al., 2014; Figure 2). In addition, the use of gene silencing methods can potentially result in concomitant silencing of closely related gene family members – perhaps to various degrees – clouding the interpretations and camouflaging the effects of downregulation of individual genes.

Figure 2. Patchy gene downregulation by RNAi. Patchy red xylem phenotype observed on trunks of CCR-deficient poplars (right) grown in a field trial in Belgium. The red xylem indicates areas of CCR downregulation. Wood from wild-type trees is whitish (left).
These issues can now be easily overcome by the use of CRISPR-based gene editing technologies that enable stable loss-of-function mutations (knock-outs) in specific target genes, allowing the dissection of the function of individual genes within families. For example, the targeting of individual 4CL gene family members in poplar showed that 4CL1 is related to lignification, whereas 4CL2 is involved in proanthocyanidin production (Zhou et al., 2015). In addition to knock-out alleles, CRISPR-based gene editing also allows to create new alleles that confer partial reduction in enzyme activity. This opens the possibility to fine-tune the level of residual enzyme activity and to bypass the yield penalty that is often observed when lignin amount drops below a threshold level. Another promising avenue for lignin engineering in forest trees made possible through CRISPR-based genome engineering is the simultaneous editing of multiple genes (allele stacking) to optimize biomass processing efficiency, as exemplified in Arabidopsis where stacking of the transaldolase (tra) and comt mutations, the c4h and comt mutations, or the 4cl and comt mutations resulted in additive and synergistic improvements in saccharification efficiency (de Vries et al., 2018). Indeed, a systems approach in P. trichocarpa predicts that the concomitant downregulation of PAL and CCoAOMT, or PAL, C3’H and CCOAOMT will substantially improve wood properties and sugar release (Wang et al., 2018).
The use of CRISPR-based genome editing in tree improvement for the pulp and paper and the bio-refinery industries, as well as for the production of platform aromatics from the hydrogenolytic breakdown of lignin, will be most valuable when this technology is strategically combined with other breeding techniques (Figure 3). Indeed, large variation in lignin amount and S/G composition already exists in natural populations of forest trees (Studer et al., 2011). Given that both traits affect the glucose release upon saccharification (Yoo et al., 2018), exploiting this genetic diversity by conventional breeding, aided by Genome Wide Association Studies (GWAS) (Porth et al., 2013; Fahrenkrog et al., 2017; Liu et al., 2018), Breeding with Rare Defective Alleles (BRDA) (Vanholme et al., 2013a) or genomic selection (Yin et al., 2010; Muchero et al., 2015; Pawar et al., 2018; Xie et al., 2018), is a valuable strategy to obtain lines that have improved wood processing efficiency. Once elite trees are obtained by these breeding methods, genetic engineering and CRISPR-based gene editing of specific genes is a very promising avenue to further improve these elite genotypes without breaking up their genetic constitution and without going through lengthy breeding cycles. Given the imminent climate crash, we have no more time to lose in adopting these new breeding techniques in our race to the biobased economy.
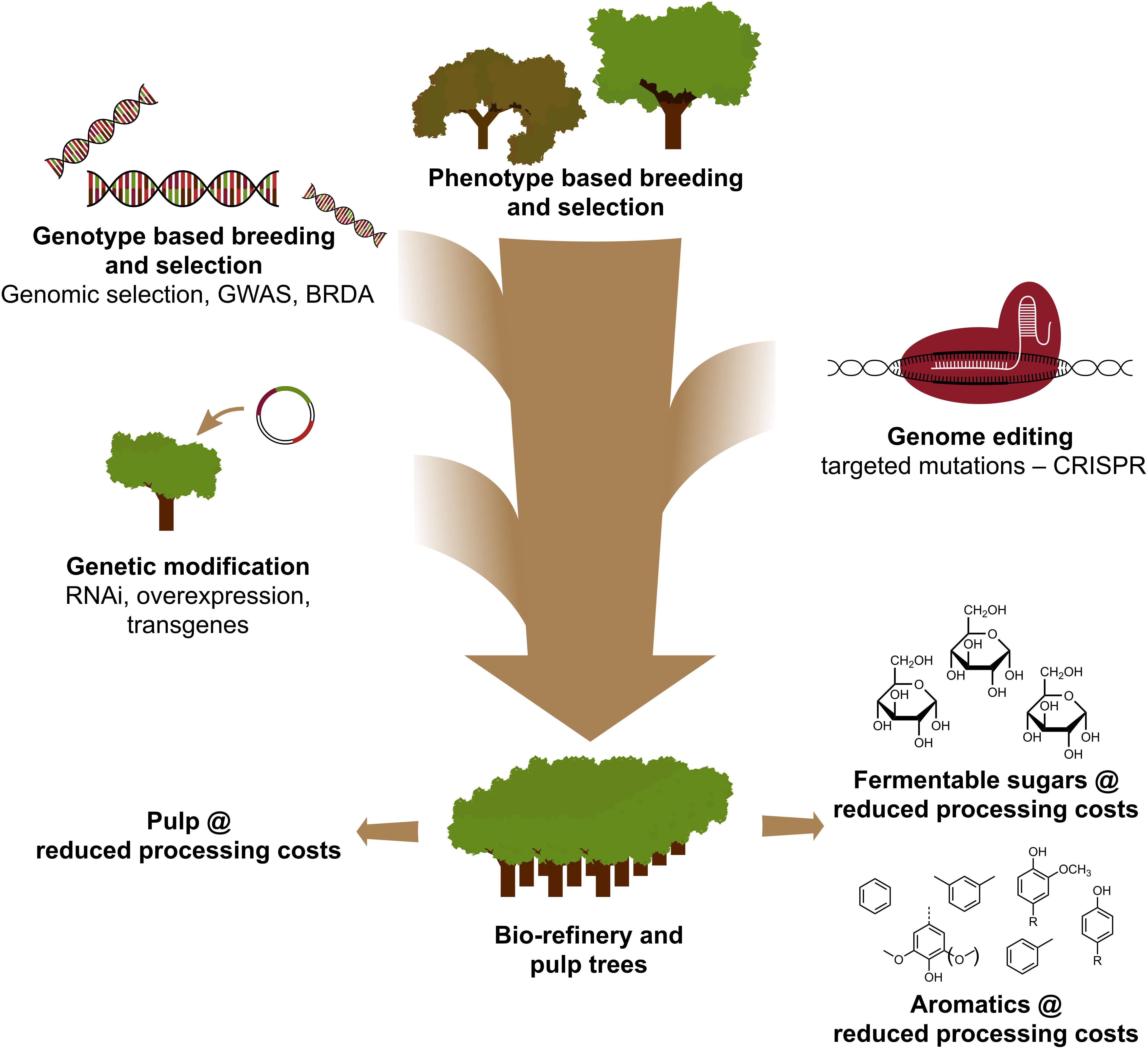
Figure 3. Genetic improvement of forest trees through a combination of breeding tools. To accelerate the genetic improvement of forest trees for pulp and biorefinery applications, classical and new breeding tools need to be smartly combined. Classical breeding involves phenotypic selection of trees for controlled crosses, followed by phenotypic selection. With the advent of genome sequence information of many forest trees, new strategies such as Genomic Selection, Genome Wide Association Studies (GWAS) and Breeding with Rare Defective Alleles (BRDA) have been developed to speed up the capture and enrichment of DNA polymorphisms associated with beneficial traits. CRISPR-based genome editing allows to modify the genome in a way that mimics natural polymorphisms. Genetic modification involves the stable integration of foreign DNA into the tree to overproduce (an) enzyme(s) or downregulate (a) gene(s). Combining the classical and New Breeding Techniques is needed to provide sufficient highly quality wood for society.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Funding
We acknowledge partial funding from the IWT-SBO project BIOLEUM (Grant No. 130039), and by SBO-FISH through the ARBOREF project. AC has received funding from the FWO and the European Union’s Horizon 2020 Research and Innovation Programme under the Marie Skłodowska-Curie Grant Agreement No. 665501. LdV was funded by the Institute for the promotion of Innovation through Science and Technology in Flanders (IWT-Vlaanderen) for a predoctoral fellowship.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Annick Bleys for preparing this manuscript for submission.
References
Awad, H., Herbette, S., Brunel, N., Tixier, A., Pilate, G., Cochard, H., et al. (2012). No trade-off between hydraulic and mechanical properties in several transgenic poplars modified for lignins metabolism. Environ. Exp. Bot. 77, 185–195. doi: 10.1016/j.envexpbot.2011.11.023
Baucher, M., Chabbert, B., Pilate, G., Vandoorsselaere, J., Tollier, M. T., Petitconil, M., et al. (1996). Red xylem and higher lignin extractability by down-regulating a cinnamyl alcohol dehydrogenase in poplar. Plant Physiol. 112, 1479–1490. doi: 10.1104/pp.112.4.1479
Beckers, B., Op De Beeck, M., Weyens, N., Boerkan, W., and Vangronsveld, J. (2017). Structural variability and niche differentiation in the rhizosphere and endosphere bacterial microbiome of field-grown poplar trees. Microbiome 5:25. doi: 10.1186/s40168-017-0241-2
Berthet, S., Demont-Caulet, N., Pollet, B., Bidzinski, P., Cézard, L., Le Bris, P., et al. (2011). Disruption of LACCASE4 and 17 results in tissue-specific alterations to lignification of Arabidopsis thaliana stems. Plant Cell 23, 1124–1137. doi: 10.1105/tpc.110.082792
Bhalla, A., Bansal, N., Pattathil, S., Li, M. Y., Shen, W., Particka, C. A., et al. (2018). Engineered lignin in poplar biomass facilitates Cu-catalyzed alkaline-oxidative pretreatment. ACS Sustain. Chem. Eng. 6, 2932–2941. doi: 10.1021/acssuschemeng.7b02067
Bhuiya, M.-W., and Liu, C.-J. (2010). Engineering monolignol 4-O-methyltransferases to modulate lignin biosynthesis. J. Biol. Chem. 285, 277–285. doi: 10.1074/jbc.M109.036673
Bjurhager, I., Olsson, A.-M., Zhang, B., Gerber, L., Kumar, M., Berglund, L. A., et al. (2010). Ultrastructure and mechanical properties of Populus wood with reduced lignin content caused by transgenic down-regulation of cinnamate 4-hydroxylase. Biomacromolecules 11, 2359–2365. doi: 10.1021/bm100487e
Boerjan, W., Ralph, J., and Baucher, M. (2003). Lignin biosynthesis. Annu. Rev. Plant Biol. 54, 519–546.
Bonawitz, N. D., and Chapple, C. (2010). The genetics of lignin biosynthesis: connecting genotype to phenotype. Annu. Rev. Genet. 44, 337–363. doi: 10.1146/annurev-genet-102209-163508
Bonawitz, N. D., Kim, J. I., Tobimatsu, Y., Ciesielski, P. N., Anderson, N. A., Ximenes, E., et al. (2014). Disruption of Mediator rescues the stunted growth of a lignin-deficient Arabidopsis mutant. Nature 509, 376–380. doi: 10.1038/nature13084
Brodeur-Campbell, S. E., Vucetich, J. A., Richter, D. L., Waite, T. A., Rosemier, J. N., and Tsai, C. J. (2006). Insect herbivory on low-lignin transgenic aspen. Environ. Entomol. 35, 1696–1701. doi: 10.1603/0046-225x(2006)35%5B1696:iholta%5D2.0.co;2
Cai, Y., Zhang, K., Kim, H., Hou, G., Zhang, X., Yang, H., et al. (2016). Enhancing digestibility and ethanol yield of Populus wood via expression of an engineered monolignol 4-O-methyltransferase. Nat. Commun. 7:11989. doi: 10.1038/ncomms11989
Cao, L., Yu, I. K. M., Liu, Y., Ruan, X., Tsang, D. C. W., Hunt, A. J., et al. (2018). Lignin valorization for the production of renewable chemicals: state-of-the-art review and future prospects. Bioresour. Technol. 269, 465–475. doi: 10.1016/j.biortech.2018.08.065
Coleman, H. D., Park, J.-Y., Nair, R., Chapple, C., and Mansfield, S. D. (2008a). Rnai-mediated suppression of p-coumaroyl-CoA 3′-hydroxylase in hybrid poplar impacts lignin deposition and soluble secondary metabolism. Proc. Natl. Acad. Sci. U.S.A. 105, 4501–4506. doi: 10.1073/pnas.0706537105
Coleman, H. D., Samuels, A. L., Guy, R. D., and Mansfield, S. D. (2008b). Perturbed lignification impacts tree growth in hybrid poplar – A function of sink strength, vascular integrity, and photosynthetic assimilation. Plant Physiol. 148, 1229–1237. doi: 10.1104/pp.108.125500
Davis, R., Tao, L., Tan, E., Biddy, M., Beckham, G., Scarlata, C., et al. (2013). Process Design and Economics for the Conversion of Lignocellulosic Biomass to Hydrocarbons: Dilute-Acid and Enzymatic Deconstruction of Biomass to Sugars and Biological Conversion of Sugars to Hydrocarbons. Golden, CO: National Renewable Energy Lab.
De Meester, B., De Vries, L., Özparpucu, M., Gierlinger, N., Corneillie, S., Pallidis, A., et al. (2018). Vessel-specific reintroduction of cinnamoyl-coa reductase1 (CCR1) in dwarfed ccr1 mutants restores vessel and xylary fiber integrity and increases biomass. Plant Physiol. 176:611. doi: 10.1104/pp.17.01462
de Vries, L., Vanholme, R., Van Acker, R., De Meester, B., Sundin, L., and Boerjan, W. (2018). Stacking of a low-lignin trait with an increased guaiacyl and 5-hydroxyguaiacyl unit trait leads to additive and synergistic effects on saccharification efficiency in Arabidopsis thaliana. Biotechnol. Biofuels 11:257. doi: 10.1186/s13068-018-1257-y
Edmunds, C. W., Peralta, P., Kelley, S. S., Chiang, V. L., Sharma-Shivappa, R. R., Davis, M. F., et al. (2017). Characterization and enzymatic hydrolysis of wood from transgenic Pinus taeda engineered with syringyl lignin or reduced lignin content. Cellulose 24, 1901–1914. doi: 10.1007/s10570-017-1231-z
Eriksson, M. E., Israelsson, M., Olsson, O., and Moritz, T. (2000). Increased gibberellin biosynthesis in transgenic trees promotes growth, biomass production and xylem fiber length. Nat. Biotechnol. 18, 784–788. doi: 10.1038/77355
Fahrenkrog, A. M., Neves, L. G., Resende, M. F. R. Jr., Vazquez, A. I., De Los Campos, G., Dervinis, C., et al. (2017). Genome-wide association study reveals putative regulators of bioenergy traits in Populus deltoides. New Phytol. 213, 799–811. doi: 10.1111/nph.14154
Franke, R., Mcmichael, C. M., Meyer, K., Shirley, A. M., Cusumano, J. C., and Chapple, C. (2000). Modified lignin in tobacco and poplar plants over-expressing the Arabidopsis gene encoding ferulate 5-hydroxylase. Plant J. 22, 223–234. doi: 10.1046/j.1365-313x.2000.00727.x
Gallego-Giraldo, L., Escamilla-Trevino, L., Jackson, L. A., and Dixon, R. A. (2011). Salicylic acid mediates the reduced growth of lignin down-regulated plants. Proc. Natl. Acad. Sci. U.S.A. 108, 20814–20819. doi: 10.1073/pnas.1117873108
Halpin, C., Thain, S. C., Tilston, E. L., Guiney, E., Lapierre, C., and Hopkins, D. W. (2007). Ecological impacts of trees with modified lignin. Tree Genet. Genomes 3, 101–110. doi: 10.1007/s11295-006-0060-2
Hjalten, J., Axelsson, E. P., Julkunen-Tiitto, R., Wennstrom, A., and Pilate, G. (2013). Innate and introduced resistance traits in genetically modified aspen trees and their effect on leaf beetle feeding. PLoS One 8:8. doi: 10.1371/journal.pone.0073819
Holladay, J. E., White, J. F., Bozell, J. J., and Johnson, D. (2007). Top Value-Added Chemicals From Biomass-Volume II—Results of Screening for Potential Candidates From Biorefinery Lignin. Richland, WA: Pacific Northwest National Lab
Hu, W.-J., Harding, S. A., Lung, J., Popko, J. L., Ralph, J., Stokke, D. D., et al. (1999). Repression of lignin biosynthesis promotes cellulose accumulation and growth in transgenic trees. Nat. Biotechnol. 17, 808–812. doi: 10.1038/11758
Huntley, S. K., Ellis, D., Gilbert, M., Chapple, C., and Mansfield, S. D. (2003). Significant increases in pulping efficiency in C4H-F5H-transformed poplars: improved chemical savings and reduced environmental toxins. J. Agric. Food Chem. 51, 6178–6183. doi: 10.1021/jf034320o
Jia, C., Zhao, H., Wang, H., Xing, Z., Du, K., Song, Y., et al. (2004). Obtaining the transgenic poplars with low lignin content through down-regulation OF4CL. Chin. Sci. Bull. 49, 905–909. doi: 10.1007/bf03184009
Jouanin, L., Goujon, T., De Nadaï, V., Martin, M.-T., Mila, I., Vallet, C., et al. (2000). Lignification in transgenic poplars with extremely reduced caffeic acid O-methyltransferase activity. Plant Physiol. 123, 1363–1373.
Kim, K. H., Dutta, T., Ralph, J., Mansfield, S. D., Simmons, B. A., and Singh, S. (2017). Impact of lignin polymer backbone esters on ionic liquid pretreatment of poplar. Biotechnol. Biofuels 10:101. doi: 10.1186/s13068-017-0784-2
Lapierre, C., Jouin, D., and Monties, B. (1989). On the molecular origin of the alkali solubility of Gramineae lignins. Phytochemistry 28, 1401–1403. doi: 10.1016/s0031-9422(00)97755-0
Lapierre, C., Pollet, B., Petit-Conil, M., Toval, G., Romero, J., Pilate, G., et al. (1999). Structural alterations of lignins in transgenic poplars with depressed cinnamyl alcohol dehydrogenase or caffeic acid O-methyltransferase activity have an opposite impact on the efficiency of industrial kraft pulping. Plant Physiol. 119, 153–163.
Lapierre, C., Pollet, B., MacKay, J. J., and Sederoff, R. R. (2000). Lignin structure in a mutant pine deficient in cinnamyl alcohol dehydrogenase. J. Agric. Food Chem. 48, 2326–2331. doi: 10.1021/jf991398p
Leplé, J.-C., Dauwe, R., Morreel, K., Storme, V., Lapierre, C., Pollet, B., et al. (2007). Downregulation of cinnamoyl-coenzyme A reductase in poplar: multiple-level phenotyping reveals effects on cell wall polymer metabolism and structure. Plant Cell 19, 3669–3691. doi: 10.1105/tpc.107.054148
Li, C., Ma, X., Yu, H., Fu, Y., and Luo, K. (2018). Ectopic expression of Ptomyb74 in poplar and arabidopsis promotes secondary cell wall formation. Front. Plant Sci. 9:1262. doi: 10.3389/fpls.2018.01262
Li, C., Zhao, X., Wang, A., Huber, G. W., and Zhang, T. (2015). Catalytic transformation of lignin for the production of chemicals and fuels. Chem. Rev. 115, 11559–11624. doi: 10.1021/acs.chemrev.5b00155
Li, C. F., Wang, X. Q., Ran, L. Y., Tian, Q. Y., Fan, D., and Luo, K. M. (2015). Ptomyb92 is a transcriptional activator of the lignin biosynthetic pathway during secondary cell wall formation in Populus tomentosa. Plant Cell Physiol. 56, 2436–2446. doi: 10.1093/pcp/pcv157
Li, L., Zhou, Y., Cheng, X., Sun, J., Marita, J. M., Ralph, J., et al. (2003). Combinatorial modification of multiple lignin traits in trees through multigene cotransformation. Proc. Natl. Acad. Sci. U.S.A. 100, 4939–4944. doi: 10.1073/pnas.0831166100
Li, Y. H., Kajita, S., Kawai, S., Katayama, Y., and Morohoshi, N. (2003). Down-regulation of an anionic peroxidase in transgenic aspen and its effect on lignin characteristics. J. Plant Res. 116, 175–182. doi: 10.1007/s10265-003-0087-5
Li, M., Pu, Y., and Ragauskas, A. J. (2016). Current understanding of the correlation of lignin structure with biomass recalcitrance. Front. Chem. 4:45.
Liang, H. Y., Frost, C. J., Wei, X. P., Brown, N. R., Carlson, J. E., and Tien, M. (2008). Improved sugar release from lignocellulosic material by introducing a tyrosine-rich cell wall peptide gene in poplar. Clean-Soil Air Water 36, 662–668. doi: 10.1002/clen.200800079
Lin, C. Y., Li, Q. Z., Tunlaya-Anukit, S., Shi, R., Sun, Y. H., Wang, J. P., et al. (2016). A cell wall-bound anionic peroxidase, Ptrpo21, is involved in lignin polymerization in Populus trichocarpa. Tree Genet. Genomes 12:18. doi: 10.1021/jf502560k
Liu, J., Ye, M., Zhu, S., Jiang, L., Sang, M., Gan, J., et al. (2018). Two-stage identification of Snp effects on dynamic poplar growth. Plant J. 93, 286–296. doi: 10.1111/tpj.13777
Lu, F. C., Marita, J. M., Lapierre, C., Jouanin, L., Morreel, K., Boerjan, W., et al. (2010). Sequencing around 5-hydroxyconiferyl alcohol-derived units in caffeic acid O-methyltransferase-deficient poplar lignins. Plant Physiol. 153, 569–579. doi: 10.1104/pp.110.154278
Lu, J., Zhao, H. Y., Wei, J. H., He, Y. K., Shi, C., Wang, H. Z., et al. (2004). Lignin reduction in transgenic poplars by expressing antisense Ccoaomt gene. Progr. Natural Sci. 14, 1060–1063. doi: 10.1080/10020070412331344801
Lu, S., Li, Q., Wei, H., Chang, M.-J., Tunlaya-Anukit, S., Kim, H., et al. (2013). Ptr-miR397a is a negative regulator of laccase genes affecting lignin content in Populus trichocarpa. Proc. Natl. Acad. Sci. U.S.A. 110, 10848–10853. doi: 10.1073/pnas.1308936110
MacKay, J. J., O’Malley, D. M., Presnell, T., Booker, F. L., Campbell, M. M., Whetten, R. W., et al. (1997). Inheritance, gene expression, and lignin characterization in a mutant pine deficient in cinnamyl alcohol-dehydrogenase. Proc. Natl. Acad. Sci. 94, 8255–8260. doi: 10.1073/pnas.94.15.8255
Mahon, E. L., and Mansfield, S. D. (2018). Tailor-made trees: engineering lignin for ease of processing and tomorrow’s bioeconomy. Curr. Opin. Biotechnol. 56, 147–155. doi: 10.1016/j.copbio.2018.10.014
Mansfield, S. D., Kang, K.-Y., and Chapple, C. (2012). Designed for deconstruction - poplar trees altered in cell wall lignification improve the efficacy of bioethanol production. New Phytol. 194, 91–101. doi: 10.1111/j.1469-8137.2011.04031.x
Marchin, R. M., Stout, A. T., Davis, A. A., and King, J. S. (2017). Transgenically altered lignin biosynthesis affects photosynthesis and water relations of field-grown Populus trichocarpa. Biomass Bioenergy 98, 15–25. doi: 10.1016/j.biombioe.2017.01.013
McCann, M. C., and Carpita, N. C. (2015). Biomass recalcitrance: a multi-scale, multi-factor, and conversion-specific property. J. Exp. Bot. 66, 4109–4118. doi: 10.1093/jxb/erv267
Meyermans, H., Morreel, K., Lapierre, C., Pollet, B., De Bruyn, A., Busson, R., et al. (2000). Modifications in lignin and accumulation of phenolic glucosides in poplar xylem upon down-regulation of caffeoyl-coenzyme A O-methyltransferase, an enzyme involved in lignin biosynthesis. J. Biol. Chem. 275, 36899–36909. doi: 10.1074/jbc.m006915200
Miedes, E., Vanholme, R., Boerjan, W., and Molina, A. (2014). The role of the secondary cell wall in plant resistance to pathogens. Front. Plant Sci. 5:358. doi: 10.3389/fpls.2014.00358
Min, D., Li, Q., Jameel, H., Chiang, V., and Chang, H. M. (2012). The cellulase-mediated saccharification on wood derived from transgenic low-lignin lines of black cottonwood (Populus trichocarpa). Appl. Biochem. Biotechnol. 168, 947–955. doi: 10.1007/s12010-012-9833-2
Min, D. Y., Yang, C. M., Chiang, V., Jameel, H., and Chang, H. M. (2014). The influence of lignin-carbohydrate complexes on the cellulase-mediated saccharification II: transgenic hybrid poplars (Populus nigra L. and Populus maximowiczii A.). Fuel 116, 56–62. doi: 10.1016/j.fuel.2013.07.046
Morreel, K., Ralph, J., Lu, F., Goeminne, G., Busson, R., Herdewijn, P., et al. (2004). Phenolic profiling of caffeic acid O-methyltransferase-deficient poplar reveals novel benzodioxane oligolignols. Plant Physiol. 136, 4023–4036. doi: 10.1104/pp.104.049312
Mottiar, Y., Vanholme, R., Boerjan, W., Ralph, J., and Mansfield, S. D. (2016). Designer lignins: harnessing the plasticity of lignification. Curr. Opin. Biotechnol. 37, 190–200. doi: 10.1016/j.copbio.2015.10.009
Muchero, W., Guo, J., Difazio, S. P., Chen, J.-G., Ranjan, P., Slavov, G. T., et al. (2015). High-resolution genetic mapping of allelic variants associated with cell wall chemistry in Populus. BMC Genomics 16:24. doi: 10.1186/s12864-015-1215-z
Muro-Villanueva, F., Mao, X., and Chapple, C. (2019). Linking phenylpropanoid metabolism, lignin deposition, and plant growth inhibition. Curr. Opin. Biotechnol. 56, 202–208. doi: 10.1016/j.copbio.2018.12.008
Nishimura, H., Kamiya, A., Nagata, T., Katahira, M., and Watanabe, T. (2018). Direct evidence for alpha ether linkage between lignin and carbohydrates in wood cell walls. Sci. Rep. 8:6538. doi: 10.1038/s41598-018-24328-9
Obudulu, O., Mahler, N., Skotare, T., Bygdell, J., Abreu, I. N., Ahnlund, M., et al. (2018). A multi-omics approach reveals function of secretory carrier-associated membrane proteins in wood formation of populus trees. BMC Genomics 19:18. doi: 10.1186/s12864-017-4411-1
Pawar, P. M.-A., Schnürer, A., Mellerowicz, E. J., and Rönnberg-Wästljung, A. C. (2018). Qtl mapping of wood Ft-Ir chemotypes shows promise for improving biofuel potential in short rotation coppice willow (Salix spp.). Bioenergy Res. 11, 351–363. doi: 10.1007/s12155-018-9901-8
Pilate, G., Guiney, E., Holt, K., Petit-Conil, M., Lapierre, C., Leple, J. C., et al. (2002). Field and pulping performances of transgenic trees with altered lignification. Nat. Biotechnol. 20, 607–612. doi: 10.1038/nbt0602-607
Porth, I., Klapšte, J., Skyba, O., Hannemann, J., Mckown, A. D., Guy, R. D., et al. (2013). Genome-wide association mapping for wood characteristics in Populus identifies an array of candidate single nucleotide polymorphisms. New Phytol. 200, 710–726. doi: 10.1111/nph.12422
Ragauskas, A. J., Beckham, G. T., Biddy, M. J., Chandra, R., Chen, F., Davis, M. F., et al. (2014). Lignin valorization: improving lignin processing in the biorefinery. Science 344:1246843. doi: 10.1126/science.1246843
Ralph, J. (2006). “What makes a good monolignol substitute?,” in The Science and Lore of the Plant Cell Wall – Biosynthesis, Structure and Function, ed. T. Hayashi (Boca Raton, FL: BrownWalker Press).
Ralph, J., Akiyama, T., Coleman, H. D., and Mansfield, S. D. (2012). Effects on lignin structure of coumarate 3-hydroxylase downregulation in poplar. Bioenergy Res. 5, 1009–1019. doi: 10.1007/s12155-012-9218-y
Ralph, J., Kim, H., Lu, F., Grabber, J. H., Leplé, J.-C., Berrio-Sierra, J., et al. (2008). Identification of the structure and origin of a thioacidolysis marker compound for ferulic acid incorporation into angiosperm lignins (and an indicator for cinnamoyl CoA reductase deficiency). Plant J. 53, 368–379. doi: 10.1111/j.1365-313X.2007.03345.x
Ralph, J., Lapierre, C., and Boerjan, W. (2019). Lignin structure and its engineering. Curr. Opin. Biotechnol. 56, 240–249. doi: 10.1016/j.copbio.2019.02.019
Ralph, J., Lundquist, K., Brunow, G., Lu, F., Kim, H., Schatz, P. F., et al. (2004). Lignins: natural polymers from oxidative coupling of 4-hydroxyphenylpropanoids. Phytochem. Rev. 3, 29–60. doi: 10.1023/b:phyt.0000047809.65444.a4
Ralph, J., MacKay, J. J., Hatfield, R. D., O’Malley, D. M., Whetten, R. W., and Sederoff, R. R. (1997). Abnormal lignin in a loblolly pine mutant. Science 277, 235–239. doi: 10.1126/science.277.5323.235
Rastogi, S., and Dwivedi, U. N. (2006). Down-regulation of lignin biosynthesis in transgenic Leucaena leucocephala harboring O-methyltransferase gene. Biotechnol. Progr. 22, 609–616. doi: 10.1021/bp050206%2B
Rinaldi, R., Jastrzebski, R., Clough, M. T., Ralph, J., Kennema, M., Bruijnincx, P. C., et al. (2016). Paving the way for lignin valorisation: recent advances in bioengineering, biorefining and catalysis. Angew Chem. Int. Ed. Engl. 55, 8164–8215. doi: 10.1002/anie.201510351
Saleme, M. L. S., Cesarino, I., Vargas, L., Kim, H., Vanholme, R., Goeminne, G., et al. (2017). Silencing caffeoyl Shikimate Esterase affects lignification and improves saccharification in poplar. Plant Physiol. 175, 1040–1057. doi: 10.1104/pp.17.00920
Schutyser, W., Renders, T., Van Den Bosch, S., Koelewijn, S. F., Beckham, G. T., and Sels, B. F. (2018). Chemicals from lignin: an interplay of lignocellulose fractionation, depolymerisation, and upgrading. Chem. Soc. Rev. 47, 852–908. doi: 10.1039/c7cs00566k
Sibout, R., Le Bris, P., Legée, F., Cézard, L., Renault, H., and Lapierre, C. (2016). Structural redesigning arabidopsis lignins into alkali-soluble lignins through the expression of p-coumaroyl-CoA:monolignol transferase PMT. Plant Physiol. 170, 1358–1366. doi: 10.1104/pp.15.01877
Smith, R. A., Gonzales-Vigil, E., Karlen, S. D., Park, J. Y., Lu, F., Wilkerson, C. G., et al. (2015). Engineering monolignol p-Coumarate Conjugates into poplar and Arabidopsis lignins. Plant Physiol. 169, 2992–3001. doi: 10.1104/pp.15.00815
Stewart, J. J., Akiyama, T., Chapple, C., Ralph, J., and Mansfield, S. D. (2009). The effects on lignin structure of overexpression of ferulate 5-hydroxylase in hybrid poplar. Plant Physiol. 150, 621–635. doi: 10.1104/pp.109.137059
Stout, A. T., Davis, A. A., Domec, J. C., Yang, C. M., Shi, R., and King, J. S. (2014). Growth under field conditions affects lignin content and productivity in transgenic Populus trichocarpa with altered lignin biosynthesis. Biomass Bioenergy 68, 228–239. doi: 10.1016/j.biombioe.2014.06.008
Studer, M. H., Demartini, J. D., Davis, M. F., Sykes, R. W., Davison, B., Keller, M., et al. (2011). Lignin content in natural Populus variants affects sugar release. Proc. Natl. Acad. Sci. U.S.A. 108, 6300–6305. doi: 10.1073/pnas.1009252108
Sykes, R. W., Gjersing, E. L., Foutz, K., Rottmann, W. H., Kuhn, S. A., Foster, C. E., et al. (2015). Down-regulation of p-coumaroyl quinate/shikimate 3′-hydroxylase (C3’H) and cinnamate 4-hydroxylase (C4H) genes in the lignin biosynthetic pathway of Eucalyptus urophylla × E. grandis leads to improved sugar release. Biotechnol. Biofuels 8, 128–128. doi: 10.1186/s13068-015-0316-x
Tian, Q., Wang, X., Li, C., Lu, W., Yang, L., Jiang, Y., et al. (2013). Functional characterization of the poplar R2R3-Myb transcription factor Ptomyb216 involved in the regulation of lignin biosynthesis during wood formation. PLoS One 8:e76369. doi: 10.1371/journal.pone.0076369
Tian, X. M., Xie, J., Zhao, Y. L., Lu, H., Liu, S. C., Qu, L., et al. (2013). Sense-, antisense- and Rnai-4cl1 regulate soluble phenolic acids, cell wall components and growth in transgenic Populus tomentosa Carr. Plant Physiol. Biochem. 65, 111–119. doi: 10.1016/j.plaphy.2013.01.010
Tiimonen, H., Aronen, T., Laakso, T., Saranpää, P., Chiang, V., Ylioja, T., et al. (2005). Does lignin modification affect feeding preference or growth performance of insect herbivores in transgenic silver birch (Betula pendula Roth)? Planta 222, 699–708. doi: 10.1007/s00425-005-0002-5
Tuck, C. O., Perez, E., Horvath, I. T., Sheldon, R. A., and Poliakoff, M. (2012). Valorization of biomass: deriving more value from waste. Science 337, 695–699. doi: 10.1126/science.1218930
Upton, B. M., and Kasko, A. M. (2016). Strategies for the conversion of lignin to high-value polymeric materials: review and perspective. Chem. Rev. 116, 2275–2306. doi: 10.1021/acs.chemrev.5b00345
Van Acker, R., Déjardin, A., Desmet, S., Hoengenaert, L., Vanholme, R., Morreel, K., et al. (2017). Different routes for conifer- and sinapaldehyde and higher saccharification upon deficiency in the dehydrogenase CAD1. Plant Physiol. 175, 1018–1039. doi: 10.1104/pp.17.00834
Van Acker, R., Leplé, J.-C., Aerts, D., Storme, V., Goeminne, G., Ivens, B., et al. (2014). Improved saccharification and ethanol yield from field-grown transgenic poplar deficient in cinnamoyl-CoA reductase. Proc. Natl. Acad. Sci. U.S.A. 111:845. doi: 10.1073/pnas.1321673111
Van den Bosch, S., Schutyser, W., Vanholme, R., Driessen, T., Koelewijn, S.-F., Renders, T., et al. (2015). Reductive lignocellulose fractionation into soluble lignin-derived phenolic monomers and dimers and processable carbohydrate pulps. Energy Environ. Sci. 8, 1748–1763. doi: 10.1039/C5EE00204D
Van Doorsselaere, J., Baucher, M., Chognot, E., Chabbert, B., Tollier, M.-T., Petit-Conil, M., et al. (1995). A novel lignin in poplar trees with a reduced caffeic acid 5-hydroxyferulic acid O-methyltransferase activity. Plant J. 8, 855–864. doi: 10.1046/j.1365-313x.1995.8060855.x
Vanholme, B., Cesarino, I., Goeminne, G., Kim, H., Marroni, F., Van Acker, R., et al. (2013a). Breeding with rare defective alleles (BRDA): a natural Populus nigra Hct mutant with modified lignin as a case study. New Phytol. 198, 765–776. doi: 10.1111/nph.12179
Vanholme, B., Desmet, T., Ronsse, F., Rabaey, K., Van Breusegem, F., De Mey, M., et al. (2013b). Towards a carbon-negative sustainable bio-based economy. Front. Plant Sci. 4:174. doi: 10.3389/fpls.2013.00174
Vanholme, R., De Meester, B., Ralph, J., and Boerjan, W. (2019). Lignin biosynthesis and its integration into metabolism. Curr. Opin. Biotechnol. 56, 230–239. doi: 10.1016/j.copbio.2019.02.018
Vanholme, R., Demedts, B., Morreel, K., Ralph, J., and Boerjan, W. (2010). Lignin biosynthesis and structure. Plant Physiol. 153, 895–905.
Vanholme, R., Morreel, K., Darrah, C., Oyarce, P., Grabber, J. H., Ralph, J., et al. (2012). Metabolic engineering of novel lignin in biomass crops. New Phytol. 196, 978–1000. doi: 10.1111/j.1469-8137.2012.04337.x
Vargas, L., Cesarino, I., Vanholme, R., Voorend, W., Saleme, M. D. S., Morreel, K., et al. (2016). Improving total saccharification yield of Arabidopsis plants by vessel-specific complementation of caffeoyl shikimate esterase (cse) mutants. Biotechnol. Biofuels 9:16. doi: 10.1186/s13068-016-0551-9
Voelker, S. L., Lachenbruch, B., Meinzer, F. C., Jourdes, M., Ki, C. Y., Patten, A. M., et al. (2010). Antisense down-regulation of 4cl expression alters lignification, tree growth, and saccharification potential of field-grown poplar. Plant Physiol. 154, 874–886. doi: 10.1104/pp.110.159269
Wadenbäck, J., Von Arnold, S., Egertsdotter, U., Walter, M. H., Grima-Pettenati, J., Goffner, D., et al. (2008). Lignin biosynthesis in transgenic Norway spruce plants harboring an antisense construct for cinnamoyl CoA reductase (Ccr). Transgenic Res. 17, 379–392. doi: 10.1007/s11248-007-9113-z
Wagner, A., Donaldson, L., Kim, H., Phillips, L., Flint, H., Steward, D., et al. (2009). Suppression of 4-coumarate-CoA ligase in the coniferous gymnosperm Pinus radiata. Plant Physiol. 149, 370–383. doi: 10.1104/pp.108.125765
Wagner, A., Ralph, J., Akiyama, T., Flint, H., Phillips, L., Torr, K., et al. (2007). Exploring lignification in conifers by silencing hydroxycinnamoyl-CoA:shikimate hydroxycinnamoyltransferase in Pinus radiata. Proc. Natl. Acad. Sci. 104, 11856–11861. doi: 10.1073/pnas.0701428104
Wagner, A., Tobimatsu, Y., Goeminne, G., Phillips, L., Flint, H., Steward, D., et al. (2013). Suppression of CCR impacts metabolite profile and cell wall composition in Pinus radiata tracheary elements. Plant Mol. Biol. 81, 105–117. doi: 10.1007/s11103-012-9985-z
Wagner, A., Tobimatsu, Y., Phillips, L., Flint, H., Geddes, B., Lu, F., et al. (2015). Syringyl lignin production in conifers: proof of concept in a Pine tracheary element system. Proc. Natl. Acad. Sci. U.S.A. 112:6218. doi: 10.1073/pnas.1411926112
Wagner, A., Tobimatsu, Y., Phillips, L., Flint, H., Torr, K., Donaldson, L., et al. (2011). CCoAOMT suppression modifies lignin composition in Pinus radiata. Plant J. 67, 119–129. doi: 10.1111/j.1365-313X.2011.04580.x
Wang, H. Z., Xue, Y. X., Chen, Y. J., Li, R. F., and Wei, J. H. (2012). Lignin modification improves the biofuel production potential in transgenic Populus tomentosa. Indust. Crops Prod. 37, 170–177. doi: 10.1016/j.indcrop.2011.12.014
Wang, J. P., Matthews, M. L., Williams, C. M., Shi, R., Yang, C., Tunlaya-Anukit, S., et al. (2018). Improving wood properties for wood utilization through multi-omics integration in lignin biosynthesis. Nat. Commun. 9:1579. doi: 10.1038/s41467-018-03863-z
Wei, J. H., Wang, Y., Wang, H., Li, R., Lin, N., Ma, R., et al. (2008). Pulping performance of transgenic poplar with depressed Caffeoyl-CoA O-methyltransferase. Chin. Sci. Bull. 53, 3553–3558. doi: 10.1007/s11434-008-0477-0
Weng, J.-K., Mo, H., and Chapple, C. (2010). Over-expression of F5H in COMT-deficient Arabidopsis leads to enrichment of an unusual lignin and disruption of pollen wall formation. Plant J. 64, 898–911. doi: 10.1111/j.1365-313X.2010.04391.x
Wilkerson, C. G., Mansfield, S. D., Lu, F., Withers, S., Park, J.-Y., Karlen, S. D., et al. (2014). Monolignol ferulate transferase introduces chemically labile linkages into the lignin backbone. Science 344, 90–93. doi: 10.1126/science.1250161
Xiang, Z. Y., Sen, S. K., Min, D. Y., Savithri, D., Lu, F. C., Jameel, H., et al. (2017). Field-grown transgenic hybrid poplar with modified lignin biosynthesis to improve enzymatic saccharification efficiency. ACS Sustain. Chem. Eng. 5, 2407–2414. doi: 10.1021/acssuschemeng.6b02740
Xie, M., Muchero, W., Bryan, A. C., Yee, K., Guo, H.-B., Zhang, J., et al. (2018). A 5-enolpyruvylshikimate 3-phosphate synthase functions as a transcriptional repressor in Populus. Plant Cell 30:1645. doi: 10.1105/tpc.18.00168
Xu, C., Fu, X., Liu, R., Guo, L., Ran, L., Li, C., et al. (2017). Ptomyb170 positively regulates lignin deposition during wood formation in poplar and confers drought tolerance in transgenic Arabidopsis. Tree Physiol. 37, 1713–1726. doi: 10.1093/treephys/tpx093
Yang, L., Zhao, X., Ran, L., Li, C., Fan, D., and Luo, K. (2017). Ptomyb156 is involved in negative regulation of phenylpropanoid metabolism and secondary cell wall biosynthesis during wood formation in poplar. Sci. Rep. 7:41209. doi: 10.1038/srep41209
Yin, T., Zhang, X., Gunter, L., Priya, R., Sykes, R., Davis, M., et al. (2010). Differential detection of genetic loci underlying stem and root lignin content in Populus. PLoS One 5:e14021. doi: 10.1371/journal.pone.0014021
Yoo, C. G., Dumitrache, A., Muchero, W., Natzke, J., Akinosho, H., Li, M., et al. (2018). Significance of lignin S/G ratio in biomass recalcitrance of Populus trichocarpa variants for bioethanol production. ACS Sustain. Chem. Eng. 6, 2162–2168. doi: 10.1021/acssuschemeng.7b03586
Zeng, Y., Zhao, S., Yang, S., and Ding, S.-Y. (2014). Lignin plays a negative role in the biochemical process for producing lignocellulosic biofuels. Curr. Opin. Biotechnol. 27, 38–45. doi: 10.1016/j.copbio.2013.09.008
Zhao, Q., and Dixon, R. A. (2014). “Altering the cell wall and its impact on plant disease: from forage to bioenergy,” in Annual Review of Phytopathology, Vol. 52, ed. N. K. Vanalfen (Palo Alto: Annual Reviews).
Zhao, Q., Nakashima, J., Chen, F., Yin, Y., Fu, C., Yun, J., et al. (2013). Laccase is necessary and nonredundant with Peroxidase for lignin polymerization during vascular development in Arabidopsis. Plant Cell 25, 3976–3987. doi: 10.1105/tpc.113.117770
Zhong, R. Q., Morrison, W. H., Himmelsbach, D. S., Poole, F. L., and Ye, Z. H. (2000). Essential role of caffeoyl coenzyme A O-methyltransferase in lignin biosynthesis in woody poplar plants. Plant Physiol. 124, 563–577.
Zhou, S., Runge, T., Karlen, S. D., Ralph, J., Gonzales-Vigil, E., and Mansfield, S. D. (2017). Chemical pulping advantages of zip-lignin hybrid poplar. ChemSusChem 10, 3565–3573. doi: 10.1002/cssc.201701317
Zhou, X., Jacobs, T. B., Xue, L. J., Harding, S. A., and Tsai, C. J. (2015). Exploiting SNPS for biallelic CRISPR mutations in the outcrossing woody perennial Populus reveals 4-coumarate:CoA ligase specificity and redundancy. New Phytol. 208, 298–301. doi: 10.1111/nph.13470
Keywords: lignin, forest trees, genetic engineering, CRISPR, field trial
Citation: Chanoca A, de Vries L and Boerjan W (2019) Lignin Engineering in Forest Trees. Front. Plant Sci. 10:912. doi: 10.3389/fpls.2019.00912
Received: 29 March 2019; Accepted: 27 June 2019;
Published: 25 July 2019.
Edited by:
Chandrashekhar Pralhad Joshi, Michigan Technological University, United StatesReviewed by:
Sivakumar Pattathil, University of Georgia, United StatesKyung-Hwan Han, Michigan State University, United States
Copyright © 2019 Chanoca, de Vries and Boerjan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wout Boerjan, woboe@psb.vib-ugent.be
 Alexandra Chanoca
Alexandra Chanoca Lisanne de Vries
Lisanne de Vries Wout Boerjan
Wout Boerjan