Segmenting Purple Rapeseed Leaves in the Field from UAV RGB Imagery Using Deep Learning as an Auxiliary Means for Nitrogen Stress Detection
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Area
2.2. Field Data Acquisition
2.3. UAV Image Acquisition
2.4. Image Pre-Processing
2.5. Dataset Preparation
2.6. Network Architecture
2.7. Evaluation Metrics
2.8. Segmentation of Purple Rapeseed Leaves with Four Other Methods
3. Results
3.1. Segmentation Results Obtained with the U-Net Model
3.2. Accuracy Evaluation for the U-Net Model and Four Other Image Segmentation Methods
4. Discussion
4.1. Comparison of the Proposed Method and Four Commonly Used Methods
4.2. Influence of Image Resolution on Sample Size Selection
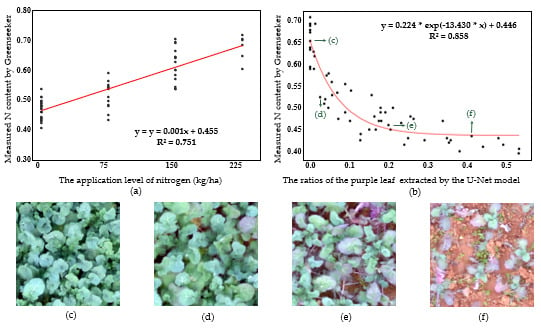
4.3. Relationship Between Nitrogen Content and Area Ratios of Purple Rapeseed Leaf
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Data Availability
Disclaimer
References
- Gitelson, A.A.; Merzlyak, M.N.; Chivkunova, O.B. Optical properties and nondestructive estimation of anthocyanin content in plant leaves. Photochem. Photobiol. 2007, 74, 38–45. [Google Scholar] [CrossRef]
- Van den Ende, W.; El-Esawe, S.K. Sucrose signaling pathways leading to fructan and anthocyanin accumulation: A dual function in abiotic and biotic stress responses? Environ. Exp. Bot. 2014, 108, 4–13. [Google Scholar] [CrossRef]
- Sakamoto, W.; Ohmori, T.; Kageyama, K.; Miyazaki, C.; Saito, A.; Murata, M.; Noda, K.; Maekawa, M. The purple leaf (Pl) locus of rice: The plw allele has a complex organization and includes two genes encoding basic helix-loop-helix proteins involved in anthocyanin biosynthesis. Plant Cell Physiol. 2001, 42, 982–991. [Google Scholar] [CrossRef] [Green Version]
- Chin, H.S.; Wu, Y.P.; Hour, A.L.; Hong, C.Y.; Lin, Y.R. Genetic and evolutionary analysis of purple leaf sheath in rice. Rice 2016, 9, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ithal, N.; Reddy, A.R. Rice flavonoid pathway genes, OsDfr and OsAns, are induced by dehydration, high salt and ABA, and contain stress responsive promoter elements that interact with the transcription activator, OsC1-MYB. Plant Sci. 2004, 166, 1505–1513. [Google Scholar] [CrossRef]
- Oren-Shamir, M.; Levi-Nissim, A. Temperature effects on the leaf pigmentation of cotinus coggygria ‘Royal Purple’. J. Hortic. Sci. 1997, 72, 425–432. [Google Scholar] [CrossRef]
- Hughes, N.M.; Lev-Yadun, S. Red/purple leaf margin coloration: Potential ecological and physiological functions. Environ. Exp. Bot. 2015, 119, 27–39. [Google Scholar] [CrossRef]
- Xiong, X.; Duan, L.; Liu, L.; Tu, H.; Yang, P.; Wu, D.; Chen, G.; Xiong, L.; Yang, W.; Liu, Q. Panicle-SEG: A robust image segmentation method for rice panicles in the field based on deep learning and superpixel optimization. Plant Methods 2017, 13, 104. [Google Scholar] [CrossRef] [Green Version]
- Tang, W.; Zhang, Y.; Zhang, D.; Yang, W.; Li, M. Corn tassel detection based on image processing. In Proceedings of the 2012 International Workshop on Image Processing and Optical Engineering, Harbin, China, 15 November 2012. [Google Scholar]
- Bai, X.D.; Cao, Z.G.; Wang, Y.; Yu, Z.H.; Zhang, X.F.; Li, C.N. Crop segmentation from images by morphology modeling in the CIE L*a*b* color space. Comput. Electron. Agric. 2013, 99, 21–34. [Google Scholar] [CrossRef]
- Tian, Y.; Li, T.; Li, C.; Piao, Z.; Sun, G.; Wang, B. Method for recognition of grape disease based on support vector machine. Trans. Chin. Soc. Agric. Eng. 2007, 23, 175–180. [Google Scholar]
- Guerrero, J.M.; Pajares, G.; Montalvo, M.; Romeo, J.; Guijarro, M. Support Vector Machines for crop/weeds identification in maize fields. Expert Syst. Appl. 2012, 39, 11149–11155. [Google Scholar] [CrossRef]
- Tatsumi, K.; Yamashiki, Y.; Torres, M.A.C.; Taipe, C.L.R. Crop classification of upland fields using Random forest of time-series Landsat 7 ETM+ data. Comput Electron. Agric. 2015, 115, 171–179. [Google Scholar] [CrossRef]
- Jeon, H.Y.; Tian, L.F.; Zhu, H. Robust crop and weed segmentation under uncontrolled outdoor illumination. Sensors 2011, 11, 6270–6283. [Google Scholar] [CrossRef]
- Liakos, K.; Busato, P.; Moshou, D.; Pearson, S.; Bochtis, D. Machine learning in agriculture: A review. Sensors 2018, 18, 2674. [Google Scholar] [CrossRef] [Green Version]
- Romera-Paredes, B.; Torr, P.H.S. Recurrent instance segmentation. In Proceedings of the 14th European Conference, Amsterdam, The Netherlands, 11–14 October 2016; pp. 312–329. [Google Scholar]
- Pound, M.P.; Atkinson, J.A.; Townsend, A.J.; Wilson, M.H.; Griffiths, M.; Jackson, A.S.; Bulat, A.; Tzimiropoulos, G.; Wells, D.M.; Murchie, E.H.; et al. Deep Machine Learning provides state-of-the-art performance in image-based plant phenotyping. Gigascience 2017, 6, 1–10. [Google Scholar] [CrossRef]
- Long, J.; Shelhamer, E.; Darrell, T. Fully convolutional networks for semantic segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2015, 79, 1337–1342. [Google Scholar]
- Sa, I.; Chen, Z.; Popović, M.; Khanna, R.; Liebisch, F.; Nieto, J.; Siegwart, R. weednet: Dense semantic weed classification using multispectral images and mav for smart farming. IEEE Robot. Autom. Lett. 2017, 3, 588–595. [Google Scholar] [CrossRef] [Green Version]
- Badrinarayanan, V.; Kendall, A.; Cipolla, R. Segnet: A deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 2481–2495. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the 18th Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015. [Google Scholar]
- Chen, L.C.; Papandreou, G.; Kokkinos, I.; Murphy, K.; Yuille, A.L. Deeplab: Semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected crfs. IEEE Trans. Pattern Anal. Mach. Intell. 2018, 40, 834–848. [Google Scholar] [CrossRef]
- Sa, I.; Popović, M.; Khanna, R.; Chen, Z.; Lottes, P.; Liebisch, F.; Nieto, J.; Stachniss, C.; Walter, A.; Siegwart, R. WeedMap: A large-scale semantic weed mapping framework using aerial multispectral imaging and deep neural network for precision farming. Remote Sens. 2018, 10, 1423. [Google Scholar] [CrossRef] [Green Version]
- Barth, R.; IJsselmuiden, J.; Hemming, J.; Van Henten, E.J. Synthetic bootstrapping of convolutional neural networks for semantic plant part segmentation. Comput. Electron. Agric. 2019, 161, 291–304. [Google Scholar] [CrossRef]
- De Brabandere, B.; Neven, D.; Van Gool, L. Semantic instance segmentation with a discriminative loss function. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 478–480. [Google Scholar]
- Milioto, A.; Lottes, P.; Stachniss, C. Real-time semantic segmentation of crop and weed for precision agriculture robots leveraging background knowledge in CNNs. In Proceedings of the IEEE International Conference on Robotics and Automation, Brisbane, Australia, 21–25 May 2018. [Google Scholar]
- Gómez-Candón, D.; De Castro, A.I.; López-Granados, F. Assessing the accuracy of mosaics from unmanned aerial vehicle (UAV) imagery for precision agriculture purposes in wheat. Precis. Agric. 2014, 15, 44–56. [Google Scholar] [CrossRef] [Green Version]
- Rango, A.; Laliberte, A.; Steele, C.; Herrick, J.E.; Bestelmeyer, B.; Schmugge, T.; Roanhorse, A.; Jenkins, V. Research article: Using unmanned aerial vehicles for rangelands: Current applications and future potentials. Environ. Pract. 2006, 8, 159–168. [Google Scholar] [CrossRef] [Green Version]
- Gong, Y.; Duan, B.; Fang, S.; Zhu, R.; Wu, X.; Ma, Y.; Peng, Y. Remote estimation of rapeseed yield with unmanned aerial vehicle (UAV) imaging and spectral mixture analysis. Plant Methods 2018, 14, 70. [Google Scholar] [CrossRef] [PubMed]
- Konoplich, G.V.; Putin, E.O.; Filchenkov, A.A. Application of deep learning to the problem of vehicle detection in UAV images. In Proceedings of the 2016 IEEE International Conference on Soft Computing and Measurements, Adygeya, Russia, 25–27 May 2016; pp. 4–6. [Google Scholar]
- Benjamin, K.; Diego, M.; Devis, T. Detecting mammals in UAV images: Best practices to address a substantially imbalanced dataset with deep learning. Remote Sens. Environ. 2018, 216, 139–153. [Google Scholar]
- Zhang, X.; Chen, G.; Wang, W.; Wang, Q.; Dai, F. Object-based land-cover supervised classification for very-high-resolution UAV images using stacked denoising autoencoders. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2017, 10, 3373–3385. [Google Scholar] [CrossRef]
- Yang, M.D.; Huang, K.S.; Kuo, Y.H.; Tsai, H.P.; Lin, L.M. Spatial and spectral hybrid image classification for rice lodging assessment through UAV imagery. Remote Sens. 2017, 9, 583. [Google Scholar] [CrossRef] [Green Version]
- Milletari, F.; Navab, N.; Ahmadi, S.A. V-Net: Fully convolutional neural networks for volumetric medical image segmentation. In Proceedings of the 14th 3D Vision, Stanford, CA, USA, 25–28 October 2016; pp. 565–571. [Google Scholar]
- Yuan, M.; Liu, Z.; Wang, F. Using the wide-range attention U-Net for road segmentation. Remote Sens. Lett. 2019, 10, 506–515. [Google Scholar] [CrossRef]
- Yao, Y.; Miao, Y.; Huang, S.; Gao, L.; Ma, X.; Zhao, G.; Jiang, R.; Chen, X.; Zhang, F.; Yu, K.; et al. Active canopy sensor-based precision N management strategy for rice. Agron. Sustain. Dev. 2012, 32, 925–933. [Google Scholar] [CrossRef] [Green Version]
- Ozer, H. Sowing date and nitrogen rate effects on growth, yield and yield components of two summer rapeseed cultivars. Eur. J. Agron. 2003, 19, 453–463. [Google Scholar] [CrossRef]
- Singh, I.; Srivastava, A.K.; Chandna, P.; Gupta, R.K. Crop sensors for efficient nitrogen management in sugarcane: Potential and constraints. Sugar Tech. 2006, 8, 299–302. [Google Scholar] [CrossRef]
- Teal, R.K.; Tubana, B.; Girma, K.; Freeman, K.W.; Arnall, D.B.; Walsh, O.; Raun, W.R. In-season prediction of corn grain yield potential using normalized difference vegetation index. Agron. J. 2006, 98, 1488–1494. [Google Scholar] [CrossRef] [Green Version]
- Ma, N.; Yuan, J.; Li, M.; Li, J.; Zhang, L.; Liu, L.; Naeem, M.S.; Zhang, C. Ideotype population exploration: Growth, photosynthesis, and yield components at different planting densities in winter oilseed rape (Brassica napus L.). PLoS ONE 2014, 9, e114232. [Google Scholar] [CrossRef]
- Wang, M.; Li, Q.; Hu, Q.; Yuan, H. A parallel interior orientation method based on lookup table for UAV images. In Proceedings of the IEEE International Conference on Spatial Data Mining and Geographical Knowledge Services, Fuzhou, China, 29 June–1 July 2011; pp. 397–400. [Google Scholar]
- Kelcey, J.; Lucieer, A. Sensor correction of a 6-band multispectral imaging sensor for UAV remote sensing. Remote Sens. 2012, 4, 1462–1493. [Google Scholar] [CrossRef] [Green Version]
- Hu, P.; Guo, W.; Chapman, S.C.; Guo, Y.; Zheng, B. Pixel size of aerial imagery constrains the applications of unmanned aerial vehicle in crop breeding. ISPRS J. Photogramm. Remote Sens. 2019, 154, 1–9. [Google Scholar] [CrossRef]
- Zhou, K.; Cheng, T.; Zhu, Y.; Cao, W.; Ustin, S.L.; Zheng, H.; Yao, X.; Tian, Y. Assessing the impact of spatial resolution on the estimation of leaf nitrogen concentration over the full season of paddy rice using near-surface imaging spectroscopy data. Front. Plant Sci. 2018, 9, 964. [Google Scholar] [CrossRef] [Green Version]
- Feng, W.; Sui, H.; Huang, W.; Xu, C.; An, K. Water body extraction from very high-resolution remote sensing imagery using deep U-Net and a superpixel-based conditional random field model. IEEE Geosci. Remote Sens. Lett. 2018, 16, 618–622. [Google Scholar] [CrossRef]
- Langner, T.; Hedström, A.; Mörwald, K.; Weghuber, D.; Forslund, A.; Bergsten, P.; Ahlström, H.; Kullberg, J. Fully convolutional networks for automated segmentation of abdominal adipose tissue depots in multicenter water–fat MRI. Magn. Reson. Med. 2019, 81, 2736–2745. [Google Scholar] [CrossRef] [Green Version]
- Kingma, D.P.; Ba, J. Adam: A method for stochastic optimization. In Proceedings of the 2014 International Conference on Learning Representations, Banff, AB, Canada, 14–16 April 2014. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Delving deep into rectifiers: Surpassing human-level performance on imageNet classification. In Proceedings of the IEEE International Conference on Computer Vision (ICCV), Santiago, Chile, 13–16 December 2015; pp. 1026–1034. [Google Scholar]
- Otsu, N. A threshold selection method from gray-level histograms. IEEE Trans. Syst. Man Cybern. 1979, 9, 62–66. [Google Scholar] [CrossRef] [Green Version]
- Puissant, A.; Rougier, S.; Stumpf, A. Object-oriented mapping of urban trees using Random Forest classifiers. Int. J. Appl. Earth Obs. Geoinf. 2014, 26, 235–245. [Google Scholar] [CrossRef]
- Kang, L.; Ye, P.; Li, Y.; Doermann, D. Convolutional neural networks for no-reference image quality assessment. In Proceedings of the 2014 IEEE Computer Vision and Pattern Recognition, Columbus, OH, USA, 23–28 June 2014. [Google Scholar]
- Camps-Valls, G.; Gómez-Chova, L.; Calpe-Maravilla, J.; Soria-Olivas, E.; Martín-Guerrero, J.D.; Moreno, J. Support vector machines for crop classification using hyperspectral data. In Proceedings of the 1st Pattern Recognition and Image Analysis, Puerto de Andratx, Mallorca, Spain, 4–6 June 2003; pp. 134–141. [Google Scholar]
- Kussul, N.; Lavreniuk, M.; Skakun, S.; Shelestov, A. Deep learning classification of land cover and crop types using remote sensing data. IEEE Geosci. Remote Sens. Lett. 2017, 14, 778–782. [Google Scholar] [CrossRef]
- Inoue, Y.; Sakaiya, E.; Zhu, Y.; Takahashi, W. Diagnostic mapping of canopy nitrogen content in rice based on hyperspectral measurements. Remote Sens. Environ. 2012, 126, 210–221. [Google Scholar] [CrossRef]
- Zhang, F.; Cui, Z.; Fan, M.; Zhang, W.; Chen, X.; Jiang, R. Integrated soil–crop system management: Reducing environmental risk while increasing crop productivity and improving nutrient use efficiency in China. J. Environ. Qual. 2011, 40, 1051–1057. [Google Scholar] [CrossRef] [PubMed]
- Wright, G.C.; Smith, C.J.; Woodroofe, M.R. The effect of irrigation and nitrogen fertilizer on rapeseed (Brassica napes) production in South-Eastern Australia: I. Growth and seed yield. Irrig. Sci. 1988, 9, 1–13. [Google Scholar] [CrossRef]
- Jia, B.; He, H.; Ma, F.; Diao, M.; Jiang, G.; Zheng, Z.; Cui, J.; Fan, H. Use of a digital camera to monitor the growth and nitrogen status of cotton. Sci. World J. 2014, 1, 1–2. [Google Scholar] [CrossRef]
- Lee, K.J.; Lee, B.W. Estimation of rice growth and nitrogen nutrition status using color digital camera image analysis. Eur. J. Agron. 2013, 48, 57–65. [Google Scholar] [CrossRef]
- Walsh, O.S.; Klatt, A.R.; Solie, J.B.; Godsey, C.B.; Raun, W.R. Use of soil moisture data for refined green seeker sensor based nitrogen recommendations in winter wheat. Precis. Agric. 2013, 14, 343–356. [Google Scholar] [CrossRef] [Green Version]
- Jay, S.; Rabatel, G.; Hadoux, X.; Moura, D.; Gorretta, N. In-field crop row phenotyping from 3D modeling performed using Structure from Motion. Comput. Electron. Agric. 2015, 110, 70–77. [Google Scholar] [CrossRef] [Green Version]
- Kaushal, S.S.; Groffman, P.M.; Band, L.E.; Elliott, E.M.; Shields, C.A.; Kendall, C. Tracking nonpoint source nitrogen pollution in human-impacted watersheds. Environ. Sci. Technol. 2011, 45, 8225–8232. [Google Scholar] [CrossRef]
- Zhang, J.; Yang, C.; Song, H.; Hoffmann, W.C.; Zhang, D.; Zhang, G. Evaluation of an airborne remote sensing platform consisting of two consumer-grade cameras for crop identification. Remote Sens. 2016, 8, 257. [Google Scholar] [CrossRef] [Green Version]









| Texture | pH | Organic Matter (g/kg) | Available N (mg/kg) | Available P (mg/kg) | Available K (mg/kg) |
|---|---|---|---|---|---|
| Silt clay loam | 6.71 | 24.16 | 133.12 | 17.16 | 145.89 |
| Patch Size | Training Set | Validation Set | Test Set | Stride |
|---|---|---|---|---|
| 64 × 64 | 62,759 | 20,920 | 20,920 | 64 |
| 128 × 128 | 25,313 | 8438 | 8438 | 128 |
| 256 × 256 | 8825 | 2941 | 2941 | 128 |
| 512 × 512 | 2841 | 947 | 947 | 128 |
| N Rate (kg/ha) | Number of Samples | Max | Min | Range | Mean | CV (%) | ANOVA Results | |
|---|---|---|---|---|---|---|---|---|
| GS values | 0 | 23 | 0.59 | 0.405 | 0.185 | 0.474d | 9.4 | p < 0.05 |
| 75 | 13 | 0.665 | 0.52 | 0.145 | 0.568c | 6.1 | ||
| 150 | 15 | 0.705 | 0.54 | 0.165 | 0.624b | 8.4 | ||
| 225 | 14 | 0.72 | 0.605 | 0.115 | 0.678a | 5.8 | ||
| Purple Leaf Ratios | 0 | 23 | 0.536 | 0.167 | 0.369 | 0.308a | 33.2 | p < 0.05 |
| 75 | 13 | 0.165 | 0.048 | 0.105 | 0.056b | 18.3 | ||
| 150 | 15 | 0.05 | 0 | 0.05 | 0.016c | 11.2 | ||
| 225 | 14 | 0.002 | 0 | 0.002 | 0.001c | 1.0 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J.; Xie, T.; Yang, C.; Song, H.; Jiang, Z.; Zhou, G.; Zhang, D.; Feng, H.; Xie, J. Segmenting Purple Rapeseed Leaves in the Field from UAV RGB Imagery Using Deep Learning as an Auxiliary Means for Nitrogen Stress Detection. Remote Sens. 2020, 12, 1403. https://doi.org/10.3390/rs12091403
Zhang J, Xie T, Yang C, Song H, Jiang Z, Zhou G, Zhang D, Feng H, Xie J. Segmenting Purple Rapeseed Leaves in the Field from UAV RGB Imagery Using Deep Learning as an Auxiliary Means for Nitrogen Stress Detection. Remote Sensing. 2020; 12(9):1403. https://doi.org/10.3390/rs12091403
Chicago/Turabian StyleZhang, Jian, Tianjin Xie, Chenghai Yang, Huaibo Song, Zhao Jiang, Guangsheng Zhou, Dongyan Zhang, Hui Feng, and Jing Xie. 2020. "Segmenting Purple Rapeseed Leaves in the Field from UAV RGB Imagery Using Deep Learning as an Auxiliary Means for Nitrogen Stress Detection" Remote Sensing 12, no. 9: 1403. https://doi.org/10.3390/rs12091403