1. Introduction
The electrocardiogram (ECG) records the tiny electrical activity produced by the heart over a period of time by placing electrodes on a patient’s body, which has become the most widely used non-invasive technique for heart disease diagnoses in the clinics. Due to the high mortality rate of heart diseases, since the last decades, ECG classification has drawn lots of researchers’ attention.
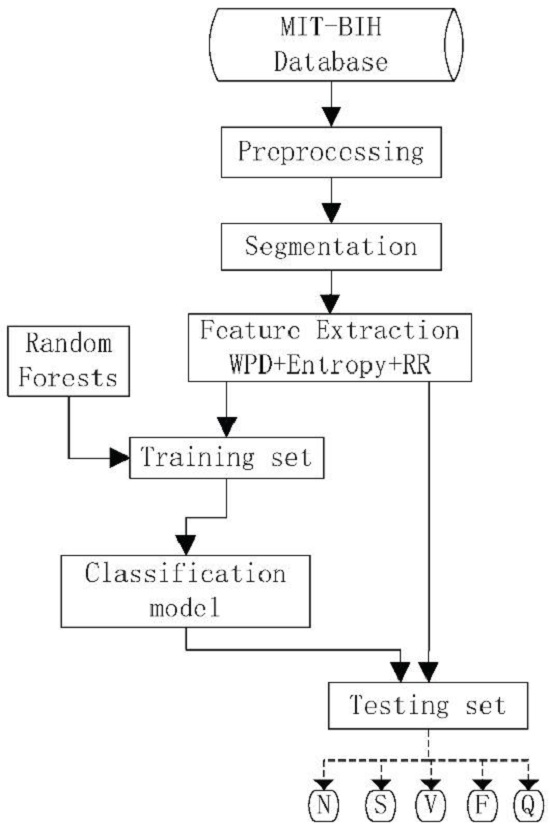
Typically, the classification of ECG signals has four phases: preprocessing, segmentation, feature extraction and classification. The preprocessing phase is mainly aimed at detecting and attenuating frequencies of the ECG signal related to artifacts, which also usually performs signal normalization and enhancement. After preprocessing, segmentation divides the signal into smaller segments, which can better express the electrical activity of the heart [
1]. Nowadays, the researchers can get good results from preprocessing and segmentation by some popular techniques or tools [
2]. Therefore, most of the literature focuses upon the last two phases.
Feature extraction plays an important role in pattern classification, especially in signal or image classification. Features can be extracted from the raw data or the transformed domain of segmented ECG signals. The simplest method of feature extraction is to extract sampled points at some frequency from an ECG signal curve [
3]. However, such a method has two drawbacks: (1) the amount of the extracted features is so huge that the efficiency of classifiers will be affected; and (2) the extracted features usually cannot reflect the intrinsic characters of the signals. Features can also be extracted using morphological and/or statistical methods from the raw signals. For example, the time between the R peaks of two heartbeats, known as the RR interval, is one of the most commonly used features. The authors used four features from the RR interval: the RR interval between the current and its predecessor heartbeats, the RR interval between the current and its successor heartbeats, the average of all the RR intervals among a full record and the average of the several neighbors’ RR intervals of the current heartbeat [
1,
4]. Independent component analysis (ICA) is another statistical method to extract ECG features. Yu et al. used ICA-based features and the RR interval to compose the feature vector. To get the ICA-based features, the authors randomly selected two sample segments and then whitened and arranged the segments into a data matrix. After that, the independent components (ICs) were calculated from the data matrix, and the original ECG signals were projected onto the bases, and the features were calculated [
5]. Later, the authors further proposed a novel IC arrangement strategy to improve the effectiveness and efficiency of ECG classification [
6]. Afkhami et al. used morphological and statistical features to train an ECG classifier [
7].
Other major feature extraction methods are to extract features from the transformed domain. Discrete cosine transform (DCT), continuous wavelet transform (CWT) and discrete wavelet transform (DWT) are commonly used transform methods. DCT expresses a finite sequence of data points in terms of a sum of cosine functions oscillating at different frequencies, which has the ability to compress signals. Khorrami et al. extracted DCT coefficients as features in ECG classification [
8]. In that paper, the authors also applied CWT and DWT to extract features for ECG classification, and compared the classification performance among DCT, CWT and DWT. Owing to its significant effectiveness for extracting discriminative features for ECG classification, wavelets were widely studied by lots of researchers. Song et al. used wavelet transform to extract 17 original input features from preprocessed signals and then reduced these to four by linear discriminant analysis (LDA), and the performance with the reduced features is better than that by principal component analysis (PCA) and even with original features [
9]. Yu and Chen used two-level DWT to decompose the signals into components in different sub-bands, and then selected three sets of statistical features of the decomposed signals, alternating current (AC) power and the instantaneous RR interval of the original signals as features [
10]. Ye et al. analysed ECG signals using morphological features (extracted by DWT and ICA) and dynamic features (RR), the feature dimensionality of the morphological ones was reduced to 26 by PCA before classification [
11]. Since the coefficients of DWT in different levels have different discrimination power, it is significant to select those that can best represent the ECG signals for classification. Daamounche et al. proposed a novel algorithm for generating the wavelet that best represents the ECG beats in terms of discrimination ability using a particle swarm optimization framework [
12]. Wavelet packet decomposition (WPD) is an extension version of DWT. Instead of decomposing approximations only in DWT, WPD decomposes both the approximations and details of the signal and hence it keeps the important information in higher-frequency components. WPD has also been applied to ECG classification. For example, the authors applied WPD to classify sleep apnea types [
13]. In another piece of literature, the authors proposed a feature extraction method based on wavelet packet of R wave window along with a strategy to select nodes in the packet tree [
14].
As for the classifiers of ECG, in theory, any multi-class classifier can be used in ECG classification. In practice, the most commonly used classifiers include support vector machine (SVM) [
15], artificial neural network (ANN), K-nearest neighbours (KNN) and decision tree (DT) [
2].
SVM is one of the most popular ECG classifiers. [
16] used a multiclass SVM with the error output codes to build a ECG classifier based on the features calculated from the wavelet coefficients. Osowski et al. [
17] presented a new approach for ECG classification by combining SVM with the features extracted by two preprocessing methods, and the results on recognizing the 13 heart rhythm types showed that the proposed method was reliable and advantageous. Mohammadzadeh et al. used SVM and the generalized discriminant analysis (GDA) feature reduction scheme to classify cardiac arrhythmia from the heart rate variability (HRV) signal [
18]. Some variations of SVM have also been applied in ECG classification, such as least square SVM [
19,
20,
21], hierarchical SVM [
22], weighted SVM [
23] and SVM combined with particle swarm optimization(PSO) [
24]. Multi-layer perception (MLP) and probabilistic neural network (PNN) are the most popular ECG classifiers associated with ANN. The authors in [
25] used the sequential forward floating search to get a feature subset and then MLP was applied to do the classification. The experimental results showed that the proposed methods exceed some previous work under the same constraints. Luz et al. compared MLP with some other classifiers on different feature sets [
1]. Alickovic and Subasi used autoregressive modeling to extract features from the de-noised signals by multiscale PCA [
26], and several classifiers including MLP were used to train models. Yu et al. used PNN to build classifiers on the combined features of RR-interval and the features by Wavelet [
10] and ICA [
5,
6], respectively. Wang and Chiang et al. pointed out that the integration of PNN with the proposed PCA and LDA feature reduction can achieve satisfactory results [
27]. Some other researchers investigated the performance of fuzzy NN [
28] and combined NN [
29] on ECG classification [
2]. Owing to the simplicity, KNN and DT were also widely applied to ECG classification. Besides the above-mentioned classifiers, some scholars also use Linear Discriminants [
4,
30,
31], Extreme Leaning Machine [
32], Optimum-path forest [
1], Active Learning [
33], and so on to build classification models.
Although so much work exists on ECG classification, it is still necessary to further explore this field. For one reason, the performance needs to be improved for modern diagnosis of heart disease. For another reason, some existing research used samples from the same patients to construct training and testing set (intra-patient scheme), which was not reasonable for practical situations because in a realistic scenario, the training samples should be from some patients and the testing should be from other patients (inter-patient scheme) [
2]. In addition, the types of cardiac arrhythmias and evaluation methods in existing research are much different, making it hard for reproducing and comparing the experiments. This can be resolved by introducing the Association for the Advancement of Medical Instrumentation (AAMI) recommendations [
34].
Entropies from WPD have been demonstrated to have a powerful ability to represent the intrinsic characteristics of electroencephalogram (EEG) signals [
35,
36]. As a robust classifier, Random Forests (RF) have been applied in many aspects, e.g., remote sensing [
37], microarray data [
38] and Alzheimer’s disease [
39]. To improve the performance of ECG classification and make the results comparable by other scholars, in this paper, we propose a new method for ECG classification using entropy on WPD and RF following the AAMI recommendations [
34] and the inter-patient scheme. The main contributions of this work are four-fold: (1) we built an ECG classification expert system with entropy on the coefficients of WPD as features and RF as a classifier; (2) we followed the AAMI recommendations and the inter-patient scheme, which made the proposed method reproducible and more practical; (3) the experimental results on the publicly accessed the MIT–BIH Arrhythmia dataset [
40] show that the proposed method is promising for ECG classification; and (4) types of entropy, mother wavelets of WPD, decomposed levels for WPD and the tree numbers of RF were discussed and the suggestions on these settings were given.
Note that the proposed method is different from the previous work [
41,
42]. Firstly, we used WPD and entropy instead of DWT coefficients to extract features. Secondly, we adopted inter-patient scheme instead of intra-patient scheme to conduct the experiments. To the best of our knowledge, it is the first time that WPE and RF are used to classify ECG following the AAMI recommendations and the inter-patient scheme.
The remainder of this paper is organized as follows.
Section 2 provides the materials and methods used, including the database, WPD, entropy, feature extraction based on WPD and entropy and the classifier RF. Experimental results are reported in
Section 3. We discuss the proposed method in
Section 4. Finally,
Section 5 concludes this paper.
4. Discussion
With the advantages of WPE and RF, we have some important issues warranting further investigation including how to choose the mother wavelet and the decomposition level for WPD, and the influence of the type of entropy and that of the number of base learners in RF. Moreover, we evaluated the efficiency of the proposed method.
To choose the best mother wavelet and the best decomposition level for WPD, we conducted experiments on six Daubechies wavelets (db1 (haar), db2, db4, db6, db8 and db10), three Coiflets wavelets (coif1, coif3 and coif5), one discrete Meyer wavelet (dmey) as well as four Biorthogonal wavelets (bior1.1, bior2.4, bior4.4 and bior6.8) with the decomposition levels ranging from 2 to 8 with an interval of 2. Here, we use Shannon entropy to extract features and 400 base learners in RF. We report the representative results in each wavelet family in
Table 7. The db4 from Daubechies wavelets family with six-level decomposition achieves the top ACC of 94.61%, slightly better than that of eight-level coif1 and six-level bioer4.4 decomposition. On average, the wavelets from the Daubechies family are the best ones for ECG classification. As far as decomposition level is concerned, a very small level can not express the signal well, while a very large one results in high-dimensional data along with many coefficients close to zero. For all the mother wavelets, the decomposition level 6 usually achieves satisfactory results.
The type of entropy is another issue in the proposed method. We adopt SE, LEE, RE as well as TE to extract features from WPT. We use real values ranging from 0.1 to 5.1 with an interval of 0.2 as the parameter
q for both RE and TE. A few typical results are shown in
Table 8. For simplicity,
Table 8 does not include the results that are very close to their neighboring ones in terms of
q. The results of LEE are slightly worse than those of others. When
q varies from 0.1 to 5.1, the performance of RE is relatively stable, with ACC varying in a narrow range of 94.28%–94.70%, indicating that RE is not sensitive to the parameter
q. The performance of TE is similar to RE with a wider range in terms of ACC. The ACC of TE reaches a minimum value of 93.8% and a maximum value of 94.64% for
,
, respectively. For RE and TE, the experimental results also vary slightly with
q in terms of SE, +P and FPR, except SE on S and F.
The number of base learners (subtrees) is also discussed here. Fixing SE and six-level db4 decomposition, we used several different numbers of base learners to validate the performance of RF. The results are shown in
Table 9. When such a number is small, e.g., 10, the performance is low. The performance increases with the increasing number to some extent. However, when the number reaches some value, e.g., 400, the performance reaches the peak. If the number continues to increase, the performance does not improve any more. We can see from
Table 9 that 400 is a good choice for the proposed methods.
We also analysed the efficiency in terms of the training and testing times with the WPE + RR features and different classifiers. For RF, we used 20, 100 and 400 as the numbers of base learners, respectively. For SVM, the time of optimizing the parameters was excluded. The results are shown in
Table 10. Since RF builds many trees in training and also votes for samples by these trees in testing, it takes more time than DT and KNN. The training and testing times increase linearly with the number of base learners. As a lazy classifier, KNN consumes the least time in training, followed by PNN and DT. The training time of PNN is only a little larger than the time to read data owing to its “one-step” training attribute. For DT, since it only needs to build a single tree, the training time is also small. The testing time of RF, DT and SVM is far less than their training time. However, for KNN and PNN, the testing time is almost 50 times and 130 times greater than the training time, respectively. In practice, the testing time plays a more important role than the training time because the training phase is usually completed with off-line data. Therefore, the time consumed by RF is acceptable.
In this approach, we calculated the entropy from the coefficients of the sixth level nodes by WPD to extract features, and the total dimension of the features is 66 including the two RR-intervals. The dimension is relatively large when compared to many existing research. We may apply PCA or best basis selection to reduce the dimension and to improve performance in future work. Since DS1 and DS2 have only eight and seven samples of class Q, respectively, showing the amounts of such classes is not representative, and none of the mentioned models can classify the samples in Q correctly, class Q should be excluded or be fused into another class in further research.