Systematic Analysis of the Pleurotus ostreatus Laccase Gene (PoLac) Family and Functional Characterization of PoLac2 Involved in the Degradation of Cotton-Straw Lignin
Abstract
:1. Introduction
2. Results
2.1. Genome-Wide Identification of Laccase Genes in P. ostreatus
2.2. Analyses of Phylogenetic Relationships, Gene Structures, and Conserved Motifs of PoLac Gene Family Members
2.3. Analyses of Multiple Sequence Alignment and Promoter Sequence
2.4. Laccase Activity of P. ostreatus and Degradation of Lignin from Cotton-Straw Medium by P. ostreatus
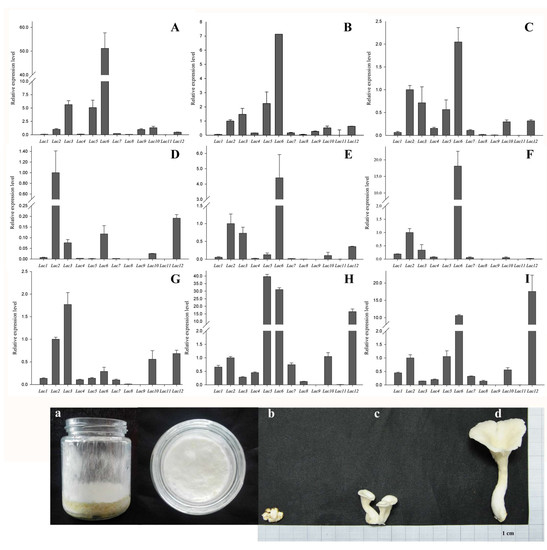
2.5. PoLac Gene Expression in Different Culture Substrates
2.6. The Lac2-Overexpressing P. ostreatus Transformants
2.7. Laccase Activity and Degradation of Lignin in Cotton-Straw Medium by Transformants
3. Discussion
4. Materials and Methods
4.1. Identification and Chromosomal Distribution of Laccase Genes in P. ostreatus
4.2. Protein Sequences and Characteristics Analysis
4.3. Phylogenetic Analysis, Gene Structure, and Conserved Motifs
4.4. Multiple Sequence Alignment and Promoter Sequence Analysis
4.5. Culture Conditions and Extraction of Enzymes
4.6. Measurement of Laccase Activity and Lignin Content
4.7. Strains Material, Total RNA Extraction, cDNA Synthesis, and qRT-PCR
4.8. Construction of Lac2 Overexpressing Plasmid Vector
4.9. Fungal Transformation
4.10. Analysis of the Transformants
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Giardina, P.; Faraco, V.; Pezzella, C.; Piscitelli, A.; Vanhulle, S.; Sannia, G. Laccases: A never-ending story. Cell Mol. Life Sci. 2010, 67, 369–385. [Google Scholar] [CrossRef] [PubMed]
- Kilaru, S.; Hoegger, P.J.; Kües, U. The laccase multi-gene family in Coprinopsis cinerea, has seventeen different members that divide into two distinct subfamilies. Curr. Genet. 2006, 50, 45–60. [Google Scholar] [CrossRef] [PubMed]
- Claus, H. Laccases and their occurrence in prokaryotes. Arch. Microbiol. 2003, 179, 145–150. [Google Scholar] [CrossRef] [PubMed]
- Cárdenas, W.; Dankert, J.R. Cresolase, catecholase and laccase activities in haemocytes of the red swamp crayfish. Fish Shellfish Immunol. 2000, 10, 33–46. [Google Scholar] [CrossRef] [PubMed]
- Zhuo, R.; Ma, L.; Fan, F.; Gong, Y.; Wan, X.; Jiang, M.; Zhang, X.; Yang, Y. Decolorization of different dyes by a newly isolated white-rot fungi strain Ganoderma sp. En3 and cloning and functional analysis of its laccase gene. J. Hazard. Mater. 2011, 192, 855–873. [Google Scholar] [CrossRef] [PubMed]
- Sigoillot, C.; Record, E.; Belle, V.; Robert, J.L.; Levasseur, A.; Punt, P.J.; van den Hondel, C.A.; Fournel, A.; Sigoillot, J.C.; Asther, M. Natural and recombinant fungal laccases for paper pulp bleaching. Appl. Microbiol. Biotechnol. 2004, 64, 346–352. [Google Scholar] [CrossRef] [PubMed]
- Ryu, S.H.; Cho, M.K.; Kim, M.; Jung, S.M.; Seo, J.H. Enhanced lignin biodegradation by a laccase-overexpressed white-rot fungus Polyporus brumalis, in the pretreatment of wood chips. Appl. Biochem. Biotechnol. 2013, 171, 1525–1534. [Google Scholar] [CrossRef] [PubMed]
- Manavalan, T.; Manavalan, A.; Heese, K. Characterization of lignocellulolytic enzymes from white-rot fungi. Curr. Microbiol. 2015, 70, 485–498. [Google Scholar] [CrossRef] [PubMed]
- Parenti, A.; Muguerza, E.; Iroz, A.R.; Omarini, A.; Conde, E.; Alfaro, M.; Castanera, R.; Santoyo, F.; Ramírez, L.; Pisabarro, A.G. Induction of laccase activity in the white rot fungus Pleurotus ostreatus using water polluted with wheat straw extracts. Bioresour. Technol. 2013, 133, 142–149. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Liu, F.; Jiang, Y.; Wu, G.; Guo, L.; Chen, R.; Chen, B.; Lu, Y.; Dai, Y.; Xie, B. The multigene family of fungal laccases and their expression in the white rot basidiomycete Flammulina velutipes. Gene 2015, 563, 142–149. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Gong, J.; Dai, W.; Kang, X.; Huang, Z.; Zhang, H.M.; Liu, W.; Liu, L.; Ma, J.; Xia, Z.; et al. Correction: The genome of Ganderma lucidum provide insights into triterpense biosynthesis and wood degradation. PLoS ONE 2012, 7, e36146. [Google Scholar] [CrossRef]
- Bao, D.; Gong, M.; Zheng, H.; Chen, M.; Zhang, L.; Wang, H.; Jiang, J.; Wu, L.; Zhu, Y.; Zhu, G.; et al. Sequencing and comparative analysis of the straw mushroom (volvariella volvacea) genome. PLoS ONE 2013, 8, e58294. [Google Scholar] [CrossRef] [PubMed]
- Soden, D.M.; Dobson, A.D. Differential regulation of laccase gene expression in Pleurotus sajor-caju. Microbiology 2001, 147, 1755–1763. [Google Scholar] [CrossRef] [PubMed]
- Alfaro, M.; Castanera, R.; Lavín, J.L.; Grigoriev, I.V.; Oguiza, J.A.; Ramírez, L.; Pisabarro, A.G. Comparative and transcriptional analysis of the predicted secretome in the lignocellulose-degrading basidiomycete fungus pleurotus ostreatus. Environ. Microbiol. 2016, 18, 4710–4726. [Google Scholar] [CrossRef] [PubMed]
- Castanera, R.; López-Varas, L.; Borgognone, A.; Labutti, K.; Lapidus, A.; Schmutz, J.; Grimwood, J.; Pérez, G.; Pisabarro, A.G.; Grigoriev, I.V.; et al. Transposable Elements versus the Fungal Genome: Impact on Whole-Genome Architecture and Transcriptional Profiles. PLoS Genet. 2016, 12, e1006108. [Google Scholar] [CrossRef] [PubMed]
- Riley, R.; Salamov, A.A.; Brown, D.W.; Nagy, L.G.; Floudas, D.; Held, B.W.; Levasseur, A.; Lombard, V.; Morin, E.; Otillar, R.; et al. Extensive sampling of basidiomycete genomes demonstrates inadequacy of the white-rot/brown-rot paradigm for wood decay fungi. Proc. Natl. Acad. Sci. USA 2014, 111, 9923–9928. [Google Scholar] [CrossRef] [PubMed]
- Pezzella, C.; Autore, F.; Giardina, P.; Piscitelli, A.; Sannia, G.; Faraco, V. The Pleurotus ostreatus laccase multi-gene family: Isolation and heterologous expression of new family members. Curr. Genet. 2009, 55, 45–57. [Google Scholar] [CrossRef] [PubMed]
- Pezzella, C.; Lettera, V.; Piscitelli, A.; Giardina, P.; Sannia, G. Transcriptional analysis of Pleurotus ostreatus laccase genes. Appl. Microbiol. Biotechnol. 2013, 97, 705–717. [Google Scholar] [CrossRef] [PubMed]
- Saparrat, M.; Balatti, P.A.; Martínez, M.J.; Jurado, M. Differential regulation of laccase gene expression in coriolopsis rigida lpsc no. 232. Fungal Biol. 2010, 114, 999–1006. [Google Scholar] [CrossRef] [PubMed]
- Arimoto, M.; Yamagishi, K.; Wang, J.; Tanaka, K.; Miyoshi, T.; Kamei, I.; Kondo, R.; Mori, T.; awagishi, H.; Hirai, H. Molecular breeding of lignin-degrading brown-rot fungus Gloeophyllum trabeum, by homologous expression of laccase gene. AMB Express. 2015, 5, 81. [Google Scholar] [CrossRef] [PubMed]
- Palmieri, G.; Giardina, P.; Bianco, C.; Fontanella, B.; Sannia, G. Copper induction of laccase isoenzymes in the ligninolytic fungus Pleurotus ostreatus. Appl. Environ. Microbiol. 2000, 66, 920–924. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Han, Y.; Li, D.; Yi, L.; Cai, Y. MYB transcription factors in chinese pear (pyrus bretschneideri rehd.): Genome-wide identification, classification, and expression profiling during fruit development. Front. Plant Sci. 2016, 7, 577. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.V.; Phale, P.S.; Durani, S.; Wangikar, P.P. Combined sequence and structure analysis of the fungal laccase family. Biotechnol. Bioeng. 2003, 83, 386–394. [Google Scholar] [CrossRef] [PubMed]
- Bertrand, T.; Jolivalt, C.; Briozzo, P.; Caminade, E.; Joly, N.; Madzak, C.; Mougin, C. Crystal structure of a four-copper laccase complexed with an arylamine: Insights into substrate recognition and correlation with kinetics. Biochemistry 2002, 41, 7325–7333. [Google Scholar] [CrossRef] [PubMed]
- Hakulinen, N.; Kiiskinen, L.L.; Kruus, K.; Saloheimo, M.; Paananen, A.; Koivula, A.; Rouvinen, J. Crystal structure of a laccase from Melanocarpus albomyces with an intact trinuclear copper site. Nat. Struct. Biol. 2002, 9, 601–605. [Google Scholar] [CrossRef] [PubMed]
- Arst, A.H.; Macdonald, D.W. Agene cluster in Aspergillus nidulans with an internally located cis-acting regulatory region. Nature 1975, 254, 26–31. [Google Scholar] [CrossRef] [PubMed]
- Varshney, U.; Jahroudi, N.; Foster, R.; Gedamu, L.; Gedamu, L. Structure, organization, and regulation of human metallothionein if gene: Differential and cell-type-specific expression in response to heavy metals and glucocorticoids. Mol. Cell Biol. 1986, 6, 26–37. [Google Scholar] [CrossRef] [PubMed]
- Mager, W.H.; De Kruijff, A.J. Stress-induced transcriptional activation. Microbiol. Rev. 1995, 59, 506–531. [Google Scholar] [CrossRef] [PubMed]
- Jarai, G.; Truong, H.N.; Daniel-Vedele, F.; Marzluf, G.A. NIT2, the nitrogen regulatory protein of Neurospora crassa, binds upstream of nia, the tomato nitrate reductase gene, in vitro. Curr. Genet. 1992, 21, 37–41. [Google Scholar] [CrossRef] [PubMed]
- Galhaup, C.; Goller, S.; Peterbauer, C.K.; Strauss, J.; Haltrich, D. Characterization of the major laccase isoenzyme from Trametes pubescens and regulation of its synthesis by metal ions. Microbiology 2002, 148, 2159–2169. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Zhou, Y.; Xiao, Y.; Xu, Z.; Bian, Y. Cloning, expression and phylogenetic analysis of a divergent laccase multigene family in Auricularia auricula-judae. Microbiol. Res. 2014, 169, 453–462. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Wu, G.; Lian, L.; Guo, L.; Wang, W.; Yang, Z.; Miao, J.; Chen, B.; Xie, B. Cloning and expression analysis of Vvlcc3, a novel and functional laccase gene possibly involved in stipe elongation. Int. J. Mol. Sci. 2015, 16, 28498–28509. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, W. The exon theory of genes. Cold Spring Harbor Symp. Quant. Biol. 1987, 52, 901–905. [Google Scholar] [CrossRef] [PubMed]
- Larrondo, L.F.; Salas, L.; Melo, F.; Vicuña, R.; Cullen, D. A novel extracellular multicopper oxidase from Phanerochaete chrysosporium with ferroxidase activity. Appl. Environ. Microbiol. 2003, 69, 6257–6262. [Google Scholar] [CrossRef] [PubMed]
- Eggert, C.; Lafayette, P.R.; Temp, U.; Eriksson, K.E.; Dean, J.F. Molecular analysis of a laccase gene from the white rot fungus Pycnoporus cinnabarinus. Appl. Environ. Microbiol. 1998, 64, 1766–1772. [Google Scholar] [PubMed]
- Jeuken, L.J.; Ubbink, M.; Bitter, J.H.; Van, V.P.; Meyer-Klaucke, W.; Canters, G.W. The structural role of the copper-coordinating and surface-exposed histidine residue in the blue copper protein azurin. J. Mol. Biol. 2000, 299, 737–755. [Google Scholar] [CrossRef] [PubMed]
- Baldrian, P.; Gabriel, J. Copper and cadmium increase laccase activity in Pleurotus ostreatus. FEMS Microbiol. Lett. 2002, 206, 69–74. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Wei, F.; Zhuo, R.; Fan, F.; Liu, H.; Zhang, C.; Ma, L.; Jiang, M.; Zhang, X. Enhancing the laccase production and laccase gene expression in the white-rot fungus Trametes velutina 5930 with great potential for biotechnological applications by different metal ions and aromatic compounds. PLoS ONE 2013, 8, e79307. [Google Scholar] [CrossRef] [PubMed]
- Álvarez, J.M.; Canessa, P.; Mancilla, R.A.; Polanco, R.; Santibáñez, P.A.; Vicuña, R. Expression of genes encoding laccase and manganese-dependent peroxidase in the fungus Ceriporiopsis subvermispora, is mediated by an ace1-like copper-fist transcription factor. Fungal Genet. Biol. 2009, 46, 104–111. [Google Scholar] [CrossRef] [PubMed]
- Lettera, V.; Piscitelli, A.; Leo, G.; Birolo, L.; Pezzella, C.; Sannia, G. Identification of a new member of Pleurotus ostreatus laccase family from mature fruiting body. Fungal Biol. 2010, 114, 724–730. [Google Scholar] [CrossRef] [PubMed]
- Fernándezfueyo, E.; Ruizdueñas, F.J.; Lópezlucendo, M.F.; Pérezboada, M.; Rencoret, J.; Gutiérrez, A.; Pisabarro, A.G.; Ramírez, L.; Martínez, A.T. A secretomic view of woody and nonwoody lignocellulose degradation by Pleurotus ostreatus. Biotechnol. Biofuels 2016, 9, 49. [Google Scholar] [CrossRef] [PubMed]
- Park, M.; Kim, M.; Kim, S.; Ha, B.; Ro, H.S. Differential expression of laccase genes in Pleurotus ostreatus and biochemical characterization of laccase isozymes produced in Pichia pastoris. Mycobiology 2015, 43, 280–287. [Google Scholar] [CrossRef] [PubMed]
- Castanera, R.; Pérez, G.; Omarini, A.; Alfaro, M.; Pisabarro, A.G.; Faraco, V.; Amore, A.; Ramíreza, L. Transcriptional and enzymatic profiling of Pleurotus ostreatus laccase genes in submerged and solid-state fermentation cultures. Appl. Environ. Microbiol. 2012, 78, 4037–4045. [Google Scholar] [CrossRef] [PubMed]
- Coconi-Linares, N.; Ortiz-Vázquez, E.; Fernández, F.; Loske, A.M.; Gómez-Lim, M.A. Recombinant expression of four oxidoreductases in Phanerochaete chrysosporium improves degradation of phenolic and non-phenolic substrates. J. Biotechnol. 2015, 209, 76–84. [Google Scholar] [CrossRef] [PubMed]
- Grinhut, T.; Salame, T.M.; Chen, Y.; Hadar, Y. Involvement of ligninolytic enzymes and fenton-like reaction in humic acid degradation by Trametes sp. Appl. Microbiol. Biotechnol. 2011, 91, 1131–1140. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Jin, J.; Liu, H.; Dong, Q.; Yan, H.; Gan, D.; Zhang, W.; Zhu, S. Genome-wide analysis of hd-zip genes in grape (Vitis vinifera). Tree Genet. Genomes 2015, 11, 827. [Google Scholar] [CrossRef]
- Blanc, G.; Wolfe, K.H. Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell 2004, 16, 1667–1678. [Google Scholar] [CrossRef] [PubMed]
- Geourjon, C.; Deléage, G. SOPMA: Significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Comput. Appl. Biosci. 1995, 11, 681–684. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Xu, Y.; Jiang, H.; Jiang, C.; Du, Y.; Gong, C.; Wang, W.; Zhu, S.; Han, G.; Cheng, B. Systematic Identification, Evolution and Expression Analysis of the Zea mays PHT1 Gene Family Reveals Several New Members Involved in Root Colonization by Arbuscular Mycorrhizal Fungi. Int. J. Mol. Sci. 2016, 17. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Elkan, C. The value of prior knowledge in discovering motifs with MEME. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1995, 3, 21–29. [Google Scholar] [PubMed]
- Xiao, Y.; Tu, X.; Wang, J.; Zhang, M.; Cheng, Q.; Zeng, W.; Shi, Y. Purification, molecular characterization and reactivity with aromatic compounds of a laccase from basidiomycete Trametes sp. strain ah28–2. Appl. Microbiol. Biotechnol. 2003, 60, 700–707. [Google Scholar] [CrossRef] [PubMed]
- Van Soest, P.J.; Robertson, J.B.; Lewis, B.A. Methods for dietary fiber, neutral detergent fiber, and nonstarch polysaccharides in relation to animal nutrition. J. Dairy Sci. 1991, 74, 3583–3597. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Castanera, R.; López-Varas, L.; Pisabarro, A.G.; Ramírez, L. Validation of Reference Genes for Transcriptional Analyses in Pleurotus ostreatus by Using Reverse Transcription-Quantitative PCR. Appl. Environ. Microbiol. 2015, 81, 4120–4129. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Liang, S.; Lei, J.; Chen, L.; Kothe, E.; Ma, A. Agrobacterium tumefaciens mediated fused egfp-hph gene expression under the control of gpd promoter in pleurotus ostreatus. Microbiol. Res. 2011, 166, 314–322. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Shi, L.; Chen, H.; Su, Y.; Zhao, M.; Ren, A.; Chen, M.; Wang, H.; Feng, Z. An efficient Agrobacterium-mediated transformation method for the edible mushroom Hypsizygus marmoreus. Microbiol. Res. 2014, 169, 741. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds not available from the authors. |








| Gene Name | Sequences ID | Number of Amino Acids (aa) | Molecular Weight (kDa) | pI | Signal Peptide Cleavage Site | Intron Number |
|---|---|---|---|---|---|---|
| PoLac1 | 1043420 | 532 | 57.57525 | 5.14 | 23–24 | 19 |
| PoLac2 | 1067328 | 522 | 57.47591 | 5.66 | 19–20 | 21 |
| PoLac3 | 1102751 | 541 | 58.86968 | 6.27 | 21–22 | 17 |
| PoLac4 | 1077328 | 522 | 57.35740 | 5.00 | 21–22 | 10 |
| PoLac5 | 1094975 | 630 | 69.36803 | 4.96 | 19–20 | 9 |
| PoLac6 | 1113032 | 533 | 57.92248 | 6.27 | 20–21 | 15 |
| PoLac7 | 1077468 | 507 | 55.62609 | 6.08 | 19–20 | 11 |
| PoLac8 | 1106925 | 534 | 58.69680 | 7.78 | 25–26 | 15 |
| PoLac9 | 1089733 | 529 | 56.58007 | 4.53 | 23–24 | 19 |
| PoLac10 | 1089723 | 533 | 56.79672 | 4.68 | 23–24 | 19 |
| PoLac11 | 1043488 | 543 | 59.41680 | 5.23 | 18–19 | 16 |
| PoLac12 | 1094965 | 513 | 55.28401 | 5.03 | 23–24 | 16 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiao, X.; Li, G.; Wang, Y.; Nie, F.; Cheng, X.; Abdullah, M.; Lin, Y.; Cai, Y. Systematic Analysis of the Pleurotus ostreatus Laccase Gene (PoLac) Family and Functional Characterization of PoLac2 Involved in the Degradation of Cotton-Straw Lignin. Molecules 2018, 23, 880. https://doi.org/10.3390/molecules23040880
Jiao X, Li G, Wang Y, Nie F, Cheng X, Abdullah M, Lin Y, Cai Y. Systematic Analysis of the Pleurotus ostreatus Laccase Gene (PoLac) Family and Functional Characterization of PoLac2 Involved in the Degradation of Cotton-Straw Lignin. Molecules. 2018; 23(4):880. https://doi.org/10.3390/molecules23040880
Chicago/Turabian StyleJiao, Xiaoyu, Guoqing Li, Yan Wang, Fan Nie, Xi Cheng, Muhammad Abdullah, Yi Lin, and Yongping Cai. 2018. "Systematic Analysis of the Pleurotus ostreatus Laccase Gene (PoLac) Family and Functional Characterization of PoLac2 Involved in the Degradation of Cotton-Straw Lignin" Molecules 23, no. 4: 880. https://doi.org/10.3390/molecules23040880