Semantic representation of scientific literature: bringing claims, contributions and named entities onto the Linked Open Data cloud
- Published
- Accepted
- Received
- Academic Editor
- Tamara Sumner
- Subject Areas
- Artificial Intelligence, Digital Libraries, Natural Language and Speech
- Keywords
- Natural language processing, Semantic web, Semantic publishing
- Copyright
- © 2015 Sateli and Witte
- Licence
- This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, reproduction and adaptation in any medium and for any purpose provided that it is properly attributed. For attribution, the original author(s), title, publication source (PeerJ Computer Science) and either DOI or URL of the article must be cited.
- Cite this article
- 2015. Semantic representation of scientific literature: bringing claims, contributions and named entities onto the Linked Open Data cloud. PeerJ Computer Science 1:e37 https://doi.org/10.7717/peerj-cs.37
Abstract
Motivation. Finding relevant scientific literature is one of the essential tasks researchers are facing on a daily basis. Digital libraries and web information retrieval techniques provide rapid access to a vast amount of scientific literature. However, no further automated support is available that would enable fine-grained access to the knowledge ‘stored’ in these documents. The emerging domain of Semantic Publishing aims at making scientific knowledge accessible to both humans and machines, by adding semantic annotations to content, such as a publication’s contributions, methods, or application domains. However, despite the promises of better knowledge access, the manual annotation of existing research literature is prohibitively expensive for wide-spread adoption. We argue that a novel combination of three distinct methods can significantly advance this vision in a fully-automated way: (i) Natural Language Processing (NLP) for Rhetorical Entity (RE) detection; (ii) Named Entity (NE) recognition based on the Linked Open Data (LOD) cloud; and (iii) automatic knowledge base construction for both NEs and REs using semantic web ontologies that interconnect entities in documents with the machine-readable LOD cloud.
Results. We present a complete workflow to transform scientific literature into a semantic knowledge base, based on the W3C standards RDF and RDFS. A text mining pipeline, implemented based on the GATE framework, automatically extracts rhetorical entities of type Claims and Contributions from full-text scientific literature. These REs are further enriched with named entities, represented as URIs to the linked open data cloud, by integrating the DBpedia Spotlight tool into our workflow. Text mining results are stored in a knowledge base through a flexible export process that provides for a dynamic mapping of semantic annotations to LOD vocabularies through rules stored in the knowledge base. We created a gold standard corpus from computer science conference proceedings and journal articles, where Claim and Contribution sentences are manually annotated with their respective types using LOD URIs. The performance of the RE detection phase is evaluated against this corpus, where it achieves an average F-measure of 0.73. We further demonstrate a number of semantic queries that show how the generated knowledge base can provide support for numerous use cases in managing scientific literature.
Availability. All software presented in this paper is available under open source licenses at http://www.semanticsoftware.info/semantic-scientific-literature-peerj-2015-supplements. Development releases of individual components are additionally available on our GitHub page at https://github.com/SemanticSoftwareLab.
Introduction
In a commentary for the Nature journal, Berners-Lee & Hendler (2001) predicted that the new semantic web technologies “may change the way scientific knowledge is produced and shared.” They envisioned the concept of “machine-understandable documents,” where machine-readable metadata is added to articles in order to explicitly mark up the data, experiments and rhetorical elements in their raw text. More than a decade later, not only is the wealth of existing publications still without annotations, but nearly all new research papers still lack semantic metadata as well. Manual efforts for adding machine-readable metadata to existing publications are simply too costly for wide-spread adoption. Hence, we investigate what kind of semantic markup can be automatically generated for research publications, in order to realize some of the envisioned benefits of semantically annotated research literature.
As part of this work, we first need to identify semantic markup that can actually help to improve specific tasks for the scientific community. A survey by Naak, Hage & Aimeur (2008) revealed that when locating papers, researchers consider two factors when assessing the relevance of a document to their information need, namely, the content and quality of the paper. They argue that a single rating value cannot represent the overall quality of a given research paper, since such a criteria can be relative to the objective of the researcher. For example, a researcher who is looking for implementation details of a specific approach is interested mostly in the Implementation section of an article and will give a higher ranking to documents with detailed technical information, rather than related documents with modest implementation details and more theoretical contributions. Therefore, a lower ranking score does not necessarily mean that the document has an overall lower (scientific) quality, but rather that its content does not satisfy the user’s current information need.
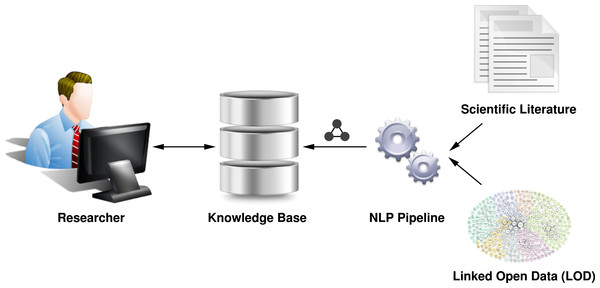
Consequently, to support users in their concrete tasks involving scientific literature, we need to go beyond standard information retrieval methods, such as keyword-based search, by taking a user’s current information need into account. Our vision (Fig. 1) is to offer support for semantically rich queries that users can ask from a knowledge base of scientific literature, including specific questions about the contributions of a publication or the discussion of specific entities, like an algorithm. For example, a user might want to ask the question “Show me all full papers from the SePublica workshops, which contain a contribution involving ‘linked data.’”
Figure 1: This diagram shows our visionary workflow to extract the knowledge contained in scientific literature by means of natural language processing (NLP), so that researchers can interact with a semantic knowledge base instead of isolated documents.
We argue that this can be achieved with a novel combination of three approaches: Natural Language Processing (NLP), Linked Open Data (LOD)-based entity detection, and semantic vocabularies for automated knowledge base construction (we discuss these methods in our ‘Background’ section below). By applying NLP techniques for rhetorical entity (RE) recognition to scientific documents, we can detect which text fragments form a rhetorical entity, like a contribution or claim. By themselves, these REs provide support for use cases such as summarization (Teufel & Moens, 2002), but cannot answer what precisely a contribution is about. We hypothesize that the named entities (NEs) present in a document (e.g., algorithms, methods, technologies) can help locate relevant publications for a user’s task. However, manually curating and updating all these possible entities for an automated NLP detection system is not a scalable solution either. Instead, we aim to leverage the Linked Open Data cloud (Heath & Bizer, 2011), which already provides a continually updated source of a wealth of knowledge across nearly every domain, with explicit and machine-readable semantics. If we can link entities detected in research papers to LOD URIs (Universal Resource Identifiers), we can semantically query a knowledge base for all papers on a specific topic (i.e., a URI), even when that topic is not mentioned literally in a text: for example, we could find a paper for the topic “linked data,” even when it only mentions “linked open data,” or even “LOD,” since they are semantically related in the DBpedia ontology (DBpedia Ontology, http://wiki.dbpedia.org/services-resources/ontology). But linked NEs alone again do not help in precisely identifying literature for a specific task: Did the paper actually make a new contribution about “linked data,” or just mention it as an application example? Our idea is that by combining the REs with the LOD NEs, we can answer questions like these in a more precise fashion than either technique alone.
To test these hypotheses, we developed a fully-automated approach that transforms publications and their NLP analysis results into a knowledge base in RDF (Resource Description Framework, http://www.w3.org/RDF) format, based on a shared vocabulary, so that they can take part in semantically rich queries and ontology-based reasoning. We evaluate the performance of this approach on several volumes of computer science conference and workshop proceedings and journal articles. Note that all queries and results shown in this paper can be verified by visiting the paper’s supplementary material webpage at http://www.semanticsoftware.info/semantic-scientific-literature-peerj-2015-supplements.
Background
Our work is based on three foundations: NLP techniques for rhetorical entity detection, named entity recognition in linked open data, and vocabularies for semantic markup of scientific documents.
Rhetorical entities
In the context of scientific literature, rhetorical entities (REs) are spans of text in a document (sentences, passages, sections, etc.), where authors convey their findings, like Claims or Arguments, to the readers. REs are usually situated in certain parts of a document, depending on their role. For example, the authors’ Claims are typically mentioned in the Abstract, Introduction or Conclusion section of a paper, and seldom in the Background. This conforms with the researchers’ habit in both reading and writing scientific articles. Indeed, according to a recent survey (Naak, Hage & Aimeur, 2008), researchers stated that they are interested in specific parts of an article when searching for literature, depending on their task at hand. Verbatim extraction of REs from text helps to efficiently allocate the attention of humans when reading a paper, as well as improving retrieval mechanisms by finding documents based on their REs (e.g., “Give me all papers with implementation details”). They can also help to narrow down the scope of subsequent knowledge extraction tasks by determining zones of text where further analysis is needed.
Existing works in automatic RE extraction are mostly based on the Rhetorical Structure Theory (RST) (Mann & Thompson, 1988) that characterizes fragments of text and the relations that hold between them, such as contrast or circumstance. Marcu (1999) developed a rhetorical parser that derives the discourse structure from unrestricted text and uses a decision tree to extract Elementary Discourse Units (EDUs) from text.
The work by Teufel (2010) identifies so-called Argumentative Zones (AZ) from scientific text as a group of sentences with the same rhetorical role. She uses statistical machine learning models and sentential features to extract AZs from a document. Teufel’s approach achieves a raw agreement of 71% with human annotations as the upper bound, using a Naïve Bayes classifier. Applications of AZs include document management and automatic summarization tasks.
In recent years, work on RE recognition has been largely limited to biomedical and chemical documents. Blake (2010) introduced the Claim Framework to differentiate levels of evidence, such as comparisons and observations, in implicit and explicit claims in biomedical domain literature. The HypothesisFinder (Malhotra et al., 2013) uses machine learning techniques to classify sentences in scientific literature in order to find speculative sentences. Combined with an ontology to find named entities in text, HypothesisFinder can establish hypothetical links between statements and their concepts in the given ontology.
The JISC-funded ART project aimed at creating an “intelligent digital library,” where the explicit semantics of scientific papers is extracted and stored using an ontology-based annotation tool. The project produced SAPIENT (Semantic Annotation of Papers: Interface & ENrichment Tool; http://www.aber.ac.uk/en/cs/research/cb/projects/art/software/), a web-based tool to help users annotate experiments in scientific papers with a set of General Specific Concepts (GSC) (Liakata & Soldatova, 2008). The development of SAPIENT was eventually succeeded by the SAPIENTA (SAPIENT Automation) tool (Liakata et al., 2012) that uses machine learning techniques to automatically annotate chemistry papers using the ART corpus as the training model. SAPIENTA’s machine learning approach has achieved an F-measure of 0.76, 0.62 and 0.53 on the automatic detection of Experiments, Background and Models (approaches) from chemistry papers, respectively.
Document markup vocabularies
An essential requirement for the semantic publishing process is the existence of controlled vocabularies that mandate the use of pre-defined terms to describe units of information with formalized, unambiguous meaning for machines. In scientific literature mining, controlled vocabularies are implemented in form of markup languages, like XML. Based on the chosen markup language, documents can be annotated for their structure and rhetorical elements, in either a manual or automatic fashion.
Structural markup
Prior to the analysis of scientific literature for their latent knowledge, we first need to provide the foundation for a common representation of documents, so that (i) the variations of their formats (e.g., HTML, PDF, LaTeX) and publisher-specific markup can be converted to one unified structure; and (ii) various segments of a document required for further processing are explicitly marked up, e.g., by separating References from the document’s main matter. A notable example is SciXML (Rupp et al., 2006), which is an XML-based markup language for domain-independent research papers. It contains a set of vocabularies that separate a document into sections that may themselves contain references, footnotes, theorems and floats, like tables and figures. SciXML also provides a stand-off1 annotation format to represent various linguistic metadata of a given document, for example, for encoding chemical terms.
The Open Annotation Model2 (OAM) (Sanderson et al., 2013) is an interoperable framework aiming towards a common specification of an annotation schema for digital resources in RDF format. The focus of the OAM is on sharing annotations for scholarly purposes with a baseline model of only three classes: a Target being annotated, a Body of information about the target, and an Annotation class that describes the relationship between the body and target, all with de-referenceable URIs.
Most of the existing annotation schemas, like SciXML, treat documents as semantically unrelated fragments of text, whereas in scientific literature this is obviously not the case: sections of a scientific article follow a logical, argumentative order (Teufel, 2010). Peroni (2012) has a similar observation and makes a distinction between XML-like languages for document markup on the one hand and semantic markup, like RDF, on the other hand. He argues that document markup languages leave the semantics of the content to the human interpretation and lack “expressiveness for the multiple and overlapping markup on the same text.” As a semantic solution, Di Iorio, Peroni & Vitali (2009) introduced the EARMARK markup metalanguage that models documents as collections of addressable text fragments and associates their content with OWL assertions to describe their structural and semantic properties. Similarly, Constantin et al. (in press) authored the DoCO3 ontology—as part of the SPAR (Semantic Publishing and Referencing) ontology family (http://www.sparontologies.net; Shotton et al., 2009)—that defines components of bibliographic documents, like figures and references, enabling their description in RDF format.
Rhetorical entity markup
In recent years, the Semantic Publishing community increasingly focused on developing vocabularies based on W3C standards, such as RDFS and OWL ontologies, for the semantic description of research publications.
SALT (Groza et al., 2007a) is a framework for the semantic annotation of scientific literature. It comprises three ontologies: a Document Ontology that defines entities like text blocks, Abstract and Title; a Rhetorical Ontology4 that defines concepts like Claims, Explanations and Results; and an Annotation Ontology that provides the means to attach syntactic and semantic markup to the document. In the early versions of the SALT framework, the embedded semantic markup was extracted from the manuscript in the compilation phase and visualized in HTML pages generated from the document metadata. The SALT framework has been extended and adapted for extracting Claims from text with the ultimate goal of creating a knowledge network from scientific publications in the KonneXSALT system (Groza et al., 2008), which provides support for (manual) identification, referencing and querying of claims in a collection of documents. Groza et al. extended their Rhetorical Ontology with concepts, such as generalizations of claims and their related text chunks, to provide for identifying claims with possible multiple representations across a dataset. They also introduced a BibTeX-like referencing system (Groza et al., 2007b) for the citation of claims that can be incorporated into the LaTeX environment using special commands, as well as queried using a web interface.
CoreSC (Liakata et al., 2010) takes on a different approach of annotating scientific documents. It treats scientific literature as a human readable representation of scientific investigations and therefore, has a vocabulary that pertains to the structure of an investigation, like Experiment or Observation. CoreSC is itself a subpart of the EXPO ontology (Soldatova et al., 2006), a comprehensive vocabulary for defining scientific experiments, like Proposition or Substrate. While ontologies like SALT or AZ-II (Teufel, Siddharthan & Batchelor, 2009) focus on the rhetorical structure of a document, ontologies like CoreSC and EXPO are used for supporting reproducibility in various domains, like chemistry or the omics sciences.
Named entity linking
An active research area in the semantic web community is concerned with recognizing entities in text and linking them to the LOD cloud (Heath & Bizer, 2011). This task is related to, but different from named entity recognition (NER) as traditionally performed in NLP in two aspects: first, only entities described on the LOD are discovered (e.g., a city name not present on an LOD source would not be detected, even if an NLP method could identify it as such) and second, each entity must be linked to a unique URI on the LOD cloud.
A well-known tool for linked NE detection is DBpedia Spotlight (Mendes et al., 2011; Daiber et al., 2013), which automatically annotates text with DBpedia resource URIs. It compares surface forms of word tokens in a text to their mentions in the DBpedia ontology. After disambiguating the sense of a word, the tool creates a link to its corresponding concept in DBpedia.
AIDA (Yosef et al., 2011) is an online tool that extracts and disambiguates NEs in a given text by calculating the prominence (frequency) and similarity of a mention to its related resources on the DBpedia, Freebase (https://www.freebase.com) and YAGO (http://www.mpi-inf.mpg.de/yago-naga/yago) ontologies.
Usbeck et al. (2014) introduced AGDISTIS, a graph-based method that is independent of the underlying LOD source and can be applied to different languages. In their evaluation, it outperformed other existing tools on several datasets.
More recently, Bontcheva et al. (2015) conducted a user study on how semantic enrichment of scientific articles can facilitate information discovery. They developed a text mining pipeline based on GATE that can process articles from the environmental science domain and link the entities in the documents to their DBpedia URI. Their goal, however, was to enrich the documents with additional metadata, such as geographical metadata, for a semantic search web service and automatically assigning a subject field to the documents from the Dublin Core (Weibel et al., 1998) ontology.
Summary
In our work, we follow an approach similar to Teufel’s in that we use NLP techniques for recognizing REs in scientific documents. However, rather than looking at documents in isolation, we aim at creating a linked data knowledge base from the documents, described with common Semantic Web vocabularies and interlinked with other LOD sources, such as DBpedia. We are not aware of existing work that combines NLP methods for RE detection with Semantic Web vocabularies in a fully-automated manner, especially in the computer science domain.
Entity linking is a highly active research area in the Semantic Web community. However, it is typically applied on general, open domain content, such as news articles or blog posts, and none of the existing datasets used for evaluation contained scientific publications. To the best of our knowledge, our work is among the first to investigate the application of entity linking on scientific documents’ LOD entities combined with rhetorical entities.
Design
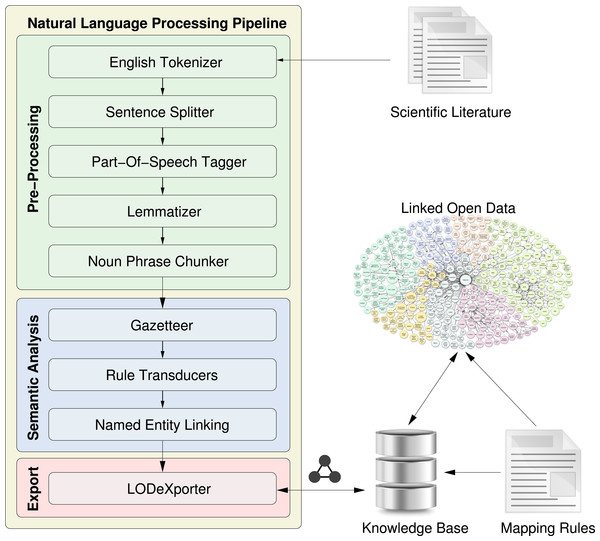
In this section, we provide a step-by-step description of our approach towards a semantic representation of scientific literature. In our system, illustrated in Fig. 2, an automatic workflow accepts scientific literature (e.g., a journal article) as input, and processes the full-text of the document to detect various syntactic and semantic entities, such as bibliographical metadata and rhetorical entities (Section ‘Automatic detection of rhetorical entities’). In addition, our approach uses NER tools to detect the topics mentioned in the document content and link them to resources on the LOD cloud (Section ‘Automatic detection of named entities’). Finally, the extracted information is stored in a semantic knowledge base (Section ‘Semantic representation of entities’), which can then be queried by humans and machines alike for their tasks.
Figure 2: A high-level overview of our workflow design, where a document is fed into an NLP pipeline that performs semantic analysis on its content and stores the extracted entities in a knowledge base, inter-linked with resources on the LOD cloud.
Automatic detection of rhetorical entities
We designed a text mining pipeline to automatically detect rhetorical entities in scientific literature, currently limited to Claims and Contributions. In our classification, Contributions are statements in a document that describe new scientific achievements attributed to its authors, such as introducing a new methodology. Claims, on the other hand, are statements by the authors that provide declarations on their contributions, such as claiming novelty or comparisons with other related works.
Our RE detection pipeline extracts such statements on a sentential level, meaning that we look at individual sentences to classify them into one of three categories: Claim, Contribution, or neither. If a chunk of text (e.g., a paragraph or section) describes a Claim or Contribution, it will be extracted as multiple, separate sentences. In our approach, we classify a document’s sentences based on the existence of several discourse elements and so-called trigger words. We adopted a rule-based approach, in which several rules are applied sequentially on a given sentence to match against its contained lexical and discourse elements. When a match is found, the rule then assigns a type, in form of a LOD URI, to the sentence under study.
Text pre-processing
As a prerequisite to the semantic analysis step, we pre-process a document’s text to covert it to a well-defined sequence of linguistically-meaningful units: words, numbers, symbols and sentences, which are passed along to the subsequent processing stages (Sateli & Witte, 2015). As part of this process, the document’s text is broken into tokens5 and lemmatized. Lemmatization is the process of finding the canonical form (lemma) of each word: e.g., “run,” “running” and “ran” all have the same root form (“run”). A Part-Of-Speech (POS) tagger then assigns a POS feature to each word token, such as noun, verb or adjective. Afterwards, determiners, adjectives and nouns are processed by a noun phrase (NP) chunker component, which groups them into NP annotations.
Gazetteering
Starting the semantic analysis phase, we perform gazetteering on the pre-processed text. Essentially, gazetteers are lists of known entities (words and phrases) that can be directly matched against the text. If a match is found, the word is tagged with its pre-defined type and later used in the rule-matching process. We manually curated multiple gazetteer lists that contain our trigger words. For example, we have gathered a list of general terms used in computer science (30 entries), such as “framework” and “approach,” as well as a comprehensive list of verbs used in the scientific argumentation context (160 entries), like “propose” and “develop,” categorized by their rhetorical functions in a text. We curated these gazetteer lists from manual inspection of the domain’s literature and Teufel’s AZ corpus (Argumentation Zoning (AZ) Corpus, http://www.cl.cam.ac.uk/~sht25/AZ_corpus.html) for rhetorical entities. In order to eliminate orthographical variations of words (e.g., plural vs. singular, past tense vs. present) the gazetteering is performed on the lemmatized text. This approach also dramatically reduces the size of the gazetteer lists, since they only need to keep the canonical form of each entry for matching against the text tokens.
Metadiscourse phrases
Detection of a rhetorical entity is performed in incremental steps: first, we detect metadiscourse elements in text, i.e., sentences where the authors describe what is being presented in the paper. Then, we classify each sentence under study based on a set of lexical and grammatical clues in the text. Metadiscourse entities often contain a discourse deixis. Deictic phrases are expressions within an utterance that refer to parts of the discourse. For example, the word “here” in “here, we describe a new methodology…” refers to the article that the user is reading. In scientific literature, deictic phrases are often used in metadiscourse phrases, such as the following examples that look for a sequence of token categories (e.g., determiner) and entries from our gazetteer (deixes are in bold):
RULEdeictic1: Determiner + Noun Phrasegazetteer
-
“This paper presents a use case of adding value to a bird observation dataset by related weather data…” (sepublica2014_paper02)
Ruledeictic2: Adverb + Punctuation
-
“Here, we demonstrate how our interpretation of NPs, named graphs, knowledge resources…” (sepublica2011_paper02)
Based on the detected deictic phrases, we annotate metadiscourse phrases in a sentence, like the following examples that are based on verbs from our gazetteer of rhetorical verbs: Rulemetadiscourse1: Deictic Phrase + Verbpresentation
-
“This paper presents a use case of adding value to a bird observation dataset by related weather data…” (sepublica2014_paper02)
Rulemetadiscourse2: Deictic Phrase + Pronoun + Verbpresentation
-
“Here, we demonstrate how our interpretation of NPs, named graphs, knowledge resources…” (sepublica2011_paper02)
Contributions
We designed hand-crafted rules to extract Contribution sentences by finding grammatical structures often observed in scientific argumentation to describe authors’ contributions. The rules look at sequences of deictic phrases, metadiscourse mentions, the rhetorical function of the verbs mentioned in the sentence and the adjacent noun phrases to classify a sentence as a Contribution, like the following example (matching string is in bold): Rulecontribution1: Metadiscourse + Noun Phrase
-
“This paper presents a use case of adding value to a bird observation dataset by related weather data…” (sepublica2014_paper02)
Rulecontribution2: Metadiscourse + Adverb + Noun Phrase
-
“Here, we demonstrate how our interpretation of NPs, named graphs, knowledge resources…” (sepublica2011_paper02)
Claims
The extraction of Claim entities is done similar to the Contribution annotations and performed based on deictic phrases detected in a text. However, here we require that the deictic phrases in Claim sentences explicitly refer to the authors’ contributions presented in the paper. Hence, we distinguish Claims from other classes in the way that the sentence containing the deictic phrase must (i) be a statement in form of a factual implication, and (ii) have a comparative voice or asserts a property of the author’s contribution, like novelty or performance:
Ruleclaim1: Metadiscourse + Determiner + Adjective + Domain Concept Trigger
-
“We built the first BauDenkMalNetz prototype using SMW [DLK+10].” (sepublica2011_paper04)
Ruleclaim2: Deictic Phrase + Verb + Domain Concept Trigger
-
“Our approach is compatible with the principles of nanopublications.” (sepublica2012_paper02)
Automatic detection of named entities
Using the rules described above, we can now find and classify REs in a scientific document. However, by using REs alone, a system is still not able to understand the topics being discussed in a document; for example, to generate a topic-focused summary. Therefore, the next step towards constructing a knowledge base of scientific literature is detecting the named entities that appear in a document. Our hypothesis here is that the extraction of named entities provides the means to represent the main topics being discussed in a paper. Therefore, the detection of the presence of such entities, along with linguistic constituents of the RE fragments, will help towards understanding the meaning of an article’s content and position of its authors regarding the detected entities, e.g., ‘enhancing algorithm A’ or ‘applying method M.’
Since the recognition of NEs varies by the functions of the field (e.g., biological terms vs. software methodologies), in lieu of developing multiple, domain-specific NER tools, we intend to reuse the LOD cloud as a structured, continually updated source of structured knowledge, by linking the surface forms of terms in a document to their corresponding resources in the LOD cloud. To further test this hypothesis, we selected the DBpedia Spotlight annotation tool described in Section ‘Named entity linking’ to automate the entity recognition task. Our goal here is to annotate the full-text of a document and then map the detected entities to the original document using the text offsets provided by Spotlight. Since we are solely interested in the named entities, we will discard any tagged entity that does not fall within a noun phrase chunk. This way, adverbs or adjectives like “here” or “successful” are filtered out and phrases like “service-oriented architecture” can be extracted as a single entity.
Semantic representation of entities
In order to transform the detected rhetorical and named entities into an interoperable and machine-understandable data structure that can be added to a semantic knowledge base, we chose to represent the extracted entities described above, as well as other metadata about each document, using the W3C standard RDF format. Therefore, each document will become a subject of a triple and all the detected entities will be attached to the document instance using custom predicates. Each entity may itself be the subject of other triples describing its semantic types and other properties, hence, creating a flexible, scalable graph of the knowledge mined from the document.
Vocabularies
As discussed in Section ‘Document markup vocabularies’, we try to reuse the existing linked open vocabularies for modeling the documents and the extracted knowledge, following the best practices for producing linked open datasets (Best Practices for Publishing Linked Data, http://www.w3.org/TR/ld-bp/). Therefore, we developed a vocabulary for scientific literature constructs partly by using existing shared vocabularies (Table 1). We chose to reuse the DoCO vocabulary for the semantic description of a document’s structure, since it covers both structural and rhetorical entities of a document through integrating the DEO (Discourse Elements Ontology, http://purl.org/spar/deo) and SALT Rhetorical Ontologies. Therefore, by using DoCO, we can describe both the structure of documents (e.g., Abstract, Title), as well as various REs types (e.g., Contributions).
We also developed our own vocabulary to describe the relations between a document and its contained entities. Our PUBlication Ontology (PUBlication Ontology (PUBO), http://lod.semanticsoftware.info/pubo/pubo.rdf) uses “pubo” as its namespace throughout this paper, and models REs as the subset of document’s sentences with a specific type, which may in turn contain a list of topics, i.e., named entities with URIs linked to their LOD resources. Figure 7 shows example RDF triples using our publication model and other shared semantic web vocabularies.
The most similar vocabulary to our PUBO vocabulary would have been the Open Annotation (OA)6 format, where each detected entity is described with a body and a target element. The former would create a URI representing the annotation (and some provenance information) and the latter provides information like the source document URL and text offsets. The generated body and target instances are then connected together using custom OA predicates. Using the OA data model, however, would lead to a ‘triple bloat’7 situation, increasing the size of knowledge base by a factor of 3–4. Moreover, the OA data model lacks an explicit representation of embedded annotations, such as the description of named entities contained within a rhetorical entity, which would require more complex and time-consuming queries to extract these facts from a knowledge base.
| Prefix | Vocabulary | URI |
|---|---|---|
| pubo | PUBlication Ontology | <http://lod.semanticsoftware.info/pubo/pubo#> |
| doco | Document Components Ontology | <http://purl.org/spar/doco> |
| sro | SALT Rhetorical Ontology | <http://salt.semanticauthoring.org/ontologies/sro#> |
| rdf | W3C RDF | <http://www.w3.org/1999/02/22-rdf-syntax-ns#> |
| rdfs | W3C RDF Schema | <http://www.w3.org/2000/01/rdf-schema#> |
| cnt | W3C Content Ontology | <http://www.w3.org/2011/content#> |
| dbpedia | DBpedia Ontology | <http://dbpedia.org/resource/> |
The entity export process
While the type of the extracted entities are decided by the rules described in ‘Automatic detection of rhetorical entities’, ideally, we still would like to have the flexibility to express the mapping of annotations to RDF triples and their inter-relations at runtime. This way, various representations of knowledge extracted from documents can be constructed based on the intended use case and customized without affecting the underlying syntactic and semantic processing components. We designed an LOD exporter component that transforms annotations in a document to RDF triples. The transformation is conducted according to a series of mapping rules. The mapping rules describe (i) the annotation type in the document and its corresponding semantic type, (ii) the annotation’s features and their corresponding semantic type, and (iii) the relations between exported triples and the type of their relation. Given the mapping rules, the exporter component then iterates over a document’s entities and exports each designated annotation as the subject of a triple, with a custom predicate and its attributes, such as its features, as the object. Table 1 summarizes the shared vocabularies that we use in the annotation export process.
Implementation
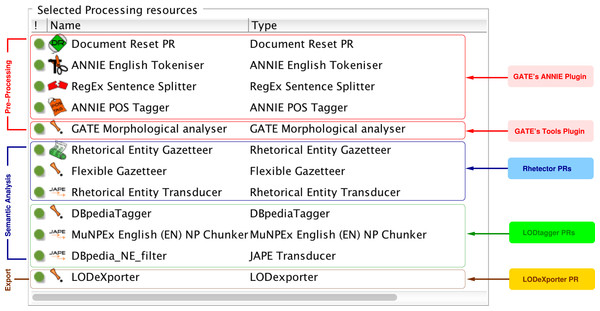
We implemented the NLP pipeline described in the ‘Design’ section based on the General Architecture for Text Engineering (GATE) (Cunningham et al., 2011),8 a robust, open-source framework for developing language engineering applications. Our pipeline is composed of several Processing Resources (PRs) that run sequentially on a given document, as shown in Fig. 3. Each processing resource can generate a new annotation or add a new feature to the annotations from upstream processing resources. In this section, we provide the implementation details of each of our pipeline’s components. Note that the materials described in this section can be found at http://www.semanticsoftware.info/semantic-scientific-literature-peerj-2015-supplements.
Figure 3: The figure shows the sequence of processing resources of our text mining pipeline that runs on a document’s text, producing various annotations, which are finally exported into a knowledge base.
Pre-processing the input documents
We use GATE’s ANNIE plugin (Cunningham et al., 2002), which offers readily available pre-processing resources to break down a document’s text into smaller units adequate for the pattern-matching rules. Specifically, we use the following processing resources provided by GATE’s ANNIE and Tools plugins:
- Document Reset PR
removes any existing annotations (e.g., from previous runs of the pipeline) from a document;
- ANNIE English Tokeniser
breaks the stream of a document’s text into tokens, classified as words, numbers or symbols;
- RegEx Sentence Splitter
uses regular expressions to detect the boundary of sentences in a document;
- ANNIE POS Tagger
adds a POS tag to each token as a new feature; and
- GATE Morphological analyser
adds the root form of each token as a new feature.
The pre-processed text is then passed onto the downstream processing resources.
Rhetector: automatic detection of rhetorical entities
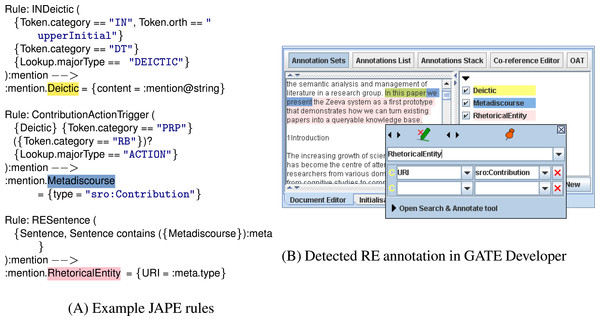
We developed Rhetector (http://www.semanticsoftware.info/rhetector) as a stand-alone GATE plugin to extract rhetorical entities from scientific literature. Rhetector has several processing resources: (i) the Rhetorical Entity Gazetteer PR that produces Lookup annotations by comparing the text tokens against its dictionary lists (domain concepts, rhetorical verbs, etc.) with the help of the Flexible Gazetteer, which looks at the root form of each token; and (ii) the Rhetorical Entity Transducer, which applies the rules described in Section ‘Automatic detection of rhetorical entities’ to sequences of Tokens and their Lookup annotations to detect rhetorical entities. The rules are implemented using GATE’s JAPE (Cunningham et al., 2011) language that provides regular expressions over document annotations, by internally compiling the rules into finite-state transducers. Every JAPE rule has a left-hand side that defines a pattern, which is matched against the text, and produces the annotation type declared on the right-hand side. Additional information are stored as features of annotations. A sequence of JAPE rules for extracting a Contribution sentence containing a metadiscourse is shown in Fig. 4.
Figure 4: The figure above shows JAPE rules (left) that are applied on a document’s text to extract a Contribution sentence.
The image on the right shows the generated annotations (Deictic, Metadiscourse and RhetoricalEntity), color-coded in GATE’s graphical user interface.LODtagger: named entity detection and grounding using DBpedia spotlight
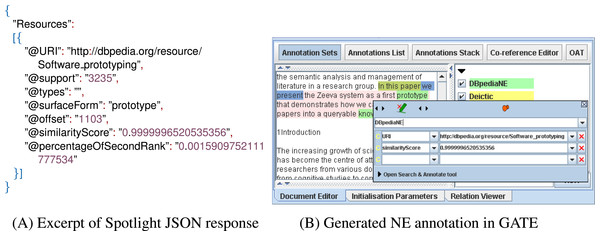
We locally installed the DBpedia Spotlight (http://spotlight.dbpedia.org) tool (Daiber et al., 2013) version 0.79 and used its RESTful annotation service to find and disambiguate named entities in our documents. To integrate the NE detection process in our semantic analysis workflow, we implemented LODtagger (http://www.semanticsoftware.info/lodtagger), a GATE plugin that acts as a wrapper for the Spotlight tool. The DBpediaTagger PR sends the full text of the document to Spotlight as an HTTP POST request and receives an array of JSON objects as the result, like the example shown in Fig. 5. The DBpediaTagger PR then parses each JSON object and adds a DBpediaLink annotation, with a DBpedia URI as its feature, to the document. To further filter the resulting entities, we align them with noun phrases (NPs), as detected by the MuNPEx NP Chunker for English.10 The aligning is performed using a JAPE rule (DBpedia_NE_filter in Fig. 3), which removes DBpediaLink annotations that are not nouns or noun phrases. Similarly, we discard NEs that include a pronoun only.
Figure 5: The figure above shows a JSON example response from Spotlight (left) and how the detected entity’s offset is used to generate a GATE annotation in the document (right).
LODeXporter: knowledge base population
We now have REs and NEs detected in the source documents, but they come in a GATE-specific data structure, i.e., GATE Annotations. In order to export them into an interoperable, queryable format, we developed LODeXporter (http://www.semanticsoftware.info/lodexporter), a GATE plugin that uses the Apache Jena (http://jena.apache.org) framework to export annotations to RDF triples, according to a set of custom mapping rules that refer to the vocabularies described in ‘Semantic representation of entities’ (cf. Table 1).
Figure 6: Example rules, expressed in RDF, declaring how GATE annotations should be mapped to RDF for knowledge base population, including the definition of LOD vocabularies to be used for the created triples.
The mapping rules themselves are also expressed using RDF and explicitly define which annotation types have to be exported and what vocabularies and relations must be used to create a new triple in the knowledge base. Using this file, each annotation becomes the subject of a new triple, with a custom predicate and its attributes, such as its features, as the object.
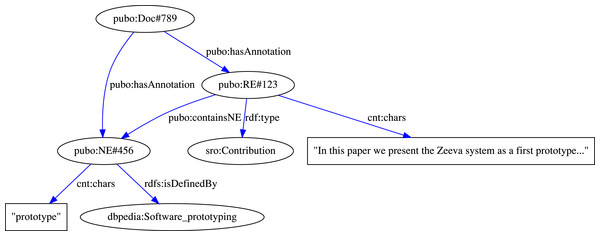
The example annotation mapping rules shown in Fig. 6 describe export specifications of RhetoricalEntity and DBpediaNE annotations in GATE documents to instances of RhetoricalElement and LinkedNamedEntity classes in the SRO and PUBO ontologies, respectively. The verbatim content of each annotation and the URI feature of each DBpediaNE is also exported using the defined predicates. Finally, using the relation mapping rule, each DBpediaNE annotation that is contained within the span of a detected RhetoricalEntity is connected to the RE instance in the knowledge base using the pubo:containsNE predicate. Ultimately, the generated RDF triples are stored in a scalable, TDB-based11 triplestore. An example RDF graph output for the mapping rules from Fig. 6 is illustrated in Fig. 7.
Figure 7: Example RDF triples generated using our publication modeling schema.
The RDF graph here represents the rhetorical and named entities annotated in a document, shown in Figs. 4 and 5, created through the mapping rules shown in Fig. 6.Evaluation
We use three open-access corpora in our experiments:
-
The SePublica corpus contains 29 documents from the proceedings of the Semantic Publishing workshops12 from 2011 to 2014.
-
PeerJCompSci is a collection of 27 open-access papers from the computer science edition of the PeerJ journal.13
-
AZ is a collection of 80 conference articles in computational linguistics, originally curated by Teufel (2010).14
The documents in these corpora are in PDF or XML formats, and range from 3 to 43 pages in various formats (ACM, LNCS, and PeerJ). We scraped the text from all files, analyzed them with our text mining pipeline described in the ‘Implementation’ section, and stored the extracted knowledge in a TDB-based triplestore.15
Quantitative analysis of the populated knowledge base
Table 2 shows the quantitative results of the populated knowledge base.16 The total number of RDF triples generated is 1,086,051. On average, the processing time of extracting REs, NEs, as well as the triplification of their relations was 5.55, 2.98 and 2.80 seconds per document for the PeerJCompSci, SePublica and AZ corpus, respectively; with the DBpedia Spotlight annotation process taking up around 60% of the processing time (running on a standard 2013 quad-core desktop PC).
For each corpus, we ran a number of queries on the knowledge base to count the occurrences of NEs and REs in the contained documents. The ‘DBpedia Named Entities (Occurrences)’ column shows the total number of NEs tagged by Spotlight, whereas the ‘DBpedia Named Entities (Distinct URIs)’ column shows the total of named entities with a unique URI. For example, if we have both “linked open data” and “LOD” tagged in a document, the total occurrence would be two, but since they are both grounded to the same URI (i.e., <dbpedia:Linked_data>), the total distinct number of NEs is one. This is particularly interesting in relation to their distribution within the documents’ rhetorical zones (column ‘Distinct DBpedia NE/RE’). As can be seen in Table 2, the number of NEs within REs are an order of a magnitude smaller than the total number of distinct named entities throughout the whole papers. This holds across the three distinct corpora we evaluated.
| Size | DBpedia named entities | Rhetorical entities | Distinct DBpediaNE/RE | |||||
|---|---|---|---|---|---|---|---|---|
| Corpus ID | Docs | Sents | Occurrences | Distinct URIs | Claims | Contributions | Claims | Contributions |
| AZ | 80 | 16,803 | 74,896 | 6,992 | 170 | 463 | 563 | 900 |
| PeerJCompSci | 27 | 15,928 | 58,808 | 8,504 | 92 | 251 | 378 | 700 |
| SePublica | 29 | 8,459 | 31,241 | 4,915 | 54 | 165 | 189 | 437 |
| Total | 136 | 41,190 | 164,945 | 14,583 | 316 | 879 | 957 | 1,643 |
This experiment shows that NEs are not evenly distributed in scientific literature. Overall, this is encouraging for our hypothesis that the combination of NEs with REs brings added value, compared to either technique alone: as mentioned in the example above, a paper could mention a topic, such as “Linked Data,” but only as part of its motivation, literature review, or future work. In this case, while the topic appears in the document, the paper does not actually contain a contribution involving linked data. Relying on standard information retrieval techniques hence results in a large amount of noise when searching for literature with a particular contribution. Semantic queries on the other hand, as we propose them here, can easily identify relevant papers in a knowledge base, as we will show in the ‘Application’ section below.
Text mining pipeline evaluation
We assessed the performance of our text mining pipeline by conducting an intrinsic evaluation i.e., comparing its precision and recall with respect to a gold standard corpus.
Gold standard corpus development and evaluation metrics
In an intrinsic evaluation scenario, the output of an NLP pipeline is directly compared with a gold standard (also known as the ground truth) to assess its performance in a task. Towards this end, we manually curated a gold standard corpus of 30 documents, where 10 papers were randomly selected from each of the three datasets described in the ‘Evaluation’ Section.
These documents were then annotated by the first author in the GATE Developer graphical user interface (Cunningham et al., 2011). Each sentence containing a rhetorical entity was manually annotated and classified as either a Claim or Contribution by adding the respective class URI from the SRO ontology as the annotation feature. The annotated SePublica papers were used during system development, whereas the annotated AZ and PeerJCompSci documents were strictly used for testing only. Table 3 shows the statistics of our gold standard corpus. Note that both the AZ and PeerjCompSci gold standard documents are available with our supplements in full-text stand-off XML format, whereas for the SePublica corpus we currently can only include our annotations, as their license does not permit redistribution.
For the evaluation, we ran our Rhetector pipeline on the evaluation corpus and computed the metrics precision (P), recall (R) and their F-measure (F-1.0), using GATE’s Corpus QA Tool (Cunningham et al., 2011). For each metric, we calculated the micro and macro average: in micro averaging, the evaluation corpus (composed of our three datasets) is treated as one large document, whereas in macro averaging, P, R and F are calculated on a per document basis, and then an average is computed (Cunningham et al., 2011).
Intrinsic evaluation results and discussion
Table 4 shows the results of our evaluation. On average, the Rhetector pipeline obtained a 0.73 F-measure on the evaluation dataset.
We gained some additional insights into the performance of Rhetector. When comparing the AZ and SePublica corpora, we can see that the pipeline achieved almost the same F-measure for roughly the same amount of text, although the two datasets are from different disciplines: SePublica documents are semantic web-related workshop papers, whereas the AZ corpus contains conference articles in computational linguistics. Another interesting observation is the robustness of Rhetector’s performance when the size of an input document (i.e., its number of tokens) increases. For example, when comparing the AZ and PeerjCompSci performance, we observed only a 0.05 difference in the pipeline’s (micro) F-measure, even though the total number of tokens to process was doubled (42,254 vs. 94,271 tokens, respectively).
An error analysis of the intrinsic evaluation results showed that the recall of our pipeline suffers when: (i) the authors’ contribution is described in passive voice and the pipeline could not attribute it to the authors, (ii) the authors used unconventional metadiscourse elements; (iii) the rhetorical entity was contained in an embedded sentence; and (iv) the sentence splitter could not find the correct sentence boundary, hence the RE span covered more than one sentence.
| Size | Annotated Rhetorical Entities | ||||
|---|---|---|---|---|---|
| Corpus ID | Documents | Sentences | Tokens | Claims | Contributions |
| AZ | 10 | 2,121 | 42,254 | 19 | 43 |
| PeerJCompSci | 10 | 5,306 | 94,271 | 36 | 62 |
| SePublica | 10 | 3,403 | 63,236 | 27 | 79 |
| Total | 30 | 10,830 | 199,761 | 82 | 184 |
| Detected Rhetorical Entities | Precision | Recall | F-1.0 | |||||
|---|---|---|---|---|---|---|---|---|
| Corpus ID | Claims | Contributions | Micro | Macro | Micro | Macro | Micro | Macro |
| AZ | 22 | 44 | 0.73 | 0.76 | 0.76 | 0.81 | 0.74 | 0.78 |
| PeerJCompSci | 32 | 86 | 0.64 | 0.70 | 0.77 | 0.72 | 0.69 | 0.69 |
| SePublica | 28 | 85 | 0.70 | 0.72 | 0.74 | 0.78 | 0.72 | 0.73 |
| Total | 82 | 215 | 0.69 | 0.73 | 0.76 | 0.77 | 0.72 | 0.73 |
Accuracy of NE grounding with Spotlight
To evaluate the accuracy of NE linking to the LOD, we randomly chose 20–50 entities per document from the SePublica corpus and manually evaluated whether they are connected to their correct sense in the DBpedia knowledge base, by inspecting their URIs through a Web browser. Out of the 120 entities manually inspected, 82 of the entities had their correct semantics in the DBpedia knowledge base. Overall, this results in 68% accuracy, which confirms our hypothesis that LOD knowledge bases are useful for the semantic description of entities in scientific documents.
Our error analysis of the detected named entities showed that Spotlight was often unable to resolve entities to their correct resource (sense) in the DBpedia knowledge base. Spotlight was also frequently unable to resolve acronyms to their full names. For example, Spotlight detected the correct sense for the term “Information Extraction,” while the term “(IE)” appearing right next to it was resolved to “Internet Explorer” instead. By design, this is exactly how the Spotlight disambiguation mechanism works: popular terms have higher chances to be connected to their surface forms. We inspected their corresponding articles on Wikipedia and discovered that the Wikipedia article on Internet Explorer is significantly longer than the Information Extraction wiki page and has 20 times more inline links, which shows its prominence in the DBpedia knowledge base, at the time of writing. Consequently, this shows that tools like Spotlight that have been trained on the general domain or news articles are biased towards topics that are more popular, which is not necessarily the best strategy for scientific publications.
Application
We published the populated knowledge base described in the previous section using the Jena Fuseki 2.017 server that provides a RESTful endpoint for SPARQL queries. We now show how the extracted knowledge can be exploited to support a user in her tasks. As a running example, let us imagine a use case: a user wants to write a literature review from a given set of documents about a specific topic.
Scenario 1. A user obtained the SePublica proceedings from the web. Before reading each article thoroughly, she would like to obtain a summary of the contributions of all articles, so she can decide which articles are relevant to her task.
Ordinarily, our user would have to read all of the retrieved documents in order to evaluate their relevance—a cumbersome and time-consuming task. However, using our approach the user can directly query for the rhetorical type that she needs from the system (note: the prefixes used in the queries in this section can be resolved using Table 1):
SELECT ?paper ?content WHERE {
?paper pubo:hasAnnotation ?rhetoricalEntity .
?rhetoricalEntity rdf:type sro:Contribution .
?rhetoricalEntity cnt:chars ?content }
ORDER BY ?paperThe system will then show the query’s results in a suitable format, like the one shown in Table 5, which dramatically reduces the amount of information that the user is exposed to, compared to a manual triage approach.
| Paper ID | Contribution |
|---|---|
| SePublica2011/paper-05.xml | “This position paper discusses how research publication would benefit of an infrastructure for evaluation entities that could be used to support documenting research efforts (e.g., in papers or blogs), analysing these efforts, and building upon them.” |
| SePublica2012/paper-03.xml | “In this paper, we describe our attempts to take a commodity publication environment, and modify it to bring in some of the formality required from academic publishing.” |
| SePublica2013/paper-05.xml | “We address the problem of identifying relations between semantic annotations and their relevance for the connectivity between related manuscripts.” |
Retrieving document sentences by their rhetorical type still returns REs that may concern entities that are irrelevant or less interesting for our user in her literature review task. Ideally, the system should return only those REs that mention user-specified topics. Since we model both the REs and NEs that appear within their boundaries, the system can allow the user to further stipulate her request. Consider the following scenario:
Scenario 2. From the set of downloaded articles, the user would like to find only those articles that have a contribution mentioning ‘linked data’.
Similar to Scenario 1, the system will answer the user’s request by executing the following query against its knowledge base:
SELECT DISTINCT ?paper ?content WHERE {
?paper pubo:hasAnnotation ?rhetoricalEntity .
?rhetoricalEntity rdf:type sro:Contribution .
?rhetoricalEntity pubo:containsNE ?ne.
?ne rdfs:isDefinedBy dbpedia:Linked_data .
?rhetoricalEntity cnt:chars ?content }
ORDER BY ?paperThe results returned by the system, partially shown in Table 6, are especially interesting. The query not only retrieved parts of articles that the user would be interested in reading, but it also inferred that “Linked Open Data,” “Linked Data” and “LOD” named entities have the same semantics, since the DBpedia knowledge base declares an <owl:sameAs> relationship between the aforementioned entities: A full-text search on the papers, on the other hand, would not have found such a semantic relation between the entities.
| Paper ID | Contribution |
|---|---|
| SePublica2012/paper-07.xml | “We present two real-life use cases in the fields of chemistry and biology and outline a general methodology for transforming research data into Linked Data.” |
| SePublica2014/paper-01.xml | “In this paper we present a vision for having such data available as Linked Open Data (LOD), and we argue that this is only possible and for the mutual benefit in cooperation between researchers and publishers.” |
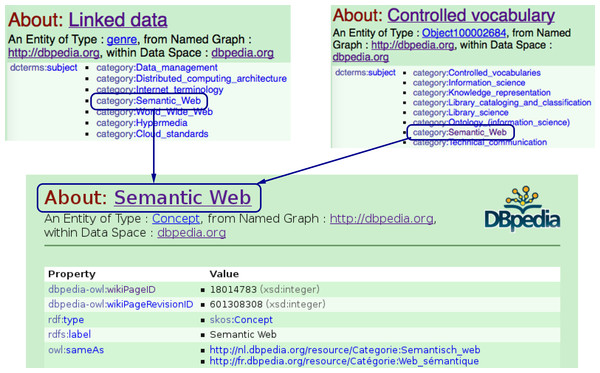
Figure 8: Finding semantically related entities in the DBpedia ontology: The Linked_data and Controlled_vocabulary entities in the DBpedia knowledge base are assumed to be semantically related to each other, since they are both contained under the same category, i.e., Semantic_Web.
So far, we showed how we can make use of the LOD-linked entities to retrieve articles of interest for a user. Note that this query returns only those articles with REs that contain an NE with a URI exactly matching that of dbpedia:Linked_data. However, by virtue of traversing the LOD cloud using an NE’s URI, we can expand the query to ask for contributions that involve dbpedia:Linked_data or any of its related subjects. In our experiment, we interpret relatedness as being under the same category in the DBpedia knowledge base (see Fig. 8). Consider the scenario below:
Scenario 3. The user would like to find only those articles that have a contribution mentioning topics related to ‘linked data’.
The system can respond to the user’s request in three steps: (i) First, through a federated query to the DBpedia knowledge base, we find the category that dbpedia:Linked_data has been assigned to—in this case, the DBpedia knowledge base returns “Semantic web,” “Data management,” and “World wide web” as the categories; (ii) Then, we retrieve all other subjects which are under the same identified categories (cf. Fig. 8); (iii) Finally, for each related entity, we look for rhetorical entities in the knowledge base that mention the related named entities within their boundaries. The semantically expanded query is shown below:
SELECT ?paper ?content WHERE {
SERVICE <http://dbpedia.org/sparql> {
dbpedia:Linked_data <http://purl.org/dc/terms/subject>
?category .
?subject <http://purl.org/dc/terms/subject> ?category . }
?paper pubo:hasAnnotation ?rhetoricalEntity .
?rhetoricalEntity rdf:type sro:Contribution .
?rhetoricalEntity pubo:containsNE ?ne.
?ne rdfs:isDefinedBy ?subject .
?rhetoricalEntity cnt:chars ?content }
ORDER BY ?paperThe system will return the results, shown in Table 7, to the user. This way, the user receives more results from the knowledge base that cover a wider range of topics semantically related to linked data, without having to explicitly define their semantic relatedness to the system. This simple example is a demonstration of how we can exploit the wealth of knowledge available in the LOD cloud. Of course, numerous other queries now become possible on scientific papers, by exploiting other linked open data sources.
| Paper ID | Contribution |
|---|---|
| SePublica2012/paper-01.xml | “In this paper, we propose a model to specify workflow-centric research objects, and show how the model can be grounded using semantic technologies and existing vocabularies, in particular the Object Reuse and Exchange (ORE) model and the Annotation Ontology (AO).” |
| SePublica2014/paper-01.xml | “In this paper we present a vision for having such data available as Linked Open Data (LOD), and we argue that this is only possible and for the mutual benefit in cooperation between researchers and publishers.” |
| SePublica2014/paper-05.xml | “In this paper we present two ontologies, i.e., BiRO and C4O, that allow users to describe bibliographic references in an accurate way, and we introduce REnhancer, a proof-of-concept implementation of a converter that takes as input a raw-text list of references and produces an RDF dataset according to the BiRO and C4O ontologies.” |
| SePublica2014/paper-07.xml | “We propose to use the CiTO ontology for describing the rhetoric of the citations (in this way we can establish a network with other works).” |
Conclusion
We all need better ways to manage the overwhelming amount of scientific literature available to us. Our approach is to create a semantic knowledge base that can supplement existing repositories, allowing users fine-grained access to documents based on querying LOD entities and their occurrence in rhetorical zones. We argue that by combining the concepts of REs and NEs, enhanced retrieval of documents becomes possible, e.g., finding all contributions on a specific topic or comparing the similarity of papers based on their REs. To demonstrate the feasibility of these ideas, we developed an NLP pipeline to fully automate the transformation of scientific documents from free-form content, read in isolation, into a queryable, semantic knowledge base. In future work, we plan to further improve both the NLP analysis and the LOD linking part of our approach. As our experiments showed, general-domain NE linking tools, like DBpedia Spotlight, are biased toward popular terms, rather than scientific entities. Here, we plan to investigate how we can adapt existing or develop new entity linking methods specifically for scientific literature. Finally, to support end users not familiar with semantic query languages, we plan to explore user interfaces and interaction patterns, e.g., based on our Zeeva semantic wiki (Sateli & Witte, 2014) system.